|
Size: 1185
Comment:
|
Size: 6077
Comment:
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 1: | Line 1: |
| == Particle Box Size and Speed == | == Particle Box Size, Reconstruction Quality and Speed == ''Warning:'' As a reminder taken from other parts of the documentation: for CTF correction to work well, it is absolutely necessary for the particle box-size to be 1.5-2x the size of the largest axis of your particle. Even if working with stain data, where accurate CTF correction may not be a priority, alignment and other routines requires ~20% padding at a bare minimum. That is, even in these cases a box size 1.2-1.3x the longest particle axis should be considered a minimum. If you go below these values, you may experience a wide range of problems. ''Reminder:'' The appropriate sampling for images for single particle reconstruction is ~2/3 Nyquist. That is, take the best resolution you hope to achieve, and divide by 3. This is close to the optimal A/pix value for your project. If your sampling is worse than this (A/pix larger), then you are not using a high enough magnification on the microscope. If your A/pix is significantly smaller than this (more than 30-50%), you should consider downsampling your data (e2proc2d.py with --meanshrink, for example). For example, if you have 2 A/pix data and are trying to achieve 20 Å resolution you may shrink the data by a factor of 2 or 3. Your refinements will run 5-10x faster, and in some cases, the results will actually be better than those with the finer sampling. ---- ''For those who don't like to read (a detailed discussion is below), here is the list of good box sizes:'' for traditional single particle analysis. Remember that for accurate CTF correction, alignment and reconstruction, the box size should normally be 1.5 - 2x the smallest box that will just contain your particle. Bold numbers also work well with shrinking by 2 or 3: * '''NEW:''' This is a list updated on 7/25/2012 for EMAN 2.06 taking into account some recent changes. Note that the new list is shorter. The detailed tables below are still based on the older analaysis. 32, 40, 44, 48, 52, '''64''', 66, '''72''', 84, 100, 104, 112, '''128''', 130, 132, 140, 150, 160, '''168''', 180, 182, 192, '''196''', 220, '''224''', '''240''', '''256''', 260, 288, 300, 320, 324, 330, 352, 360, '''384''', 416, 420, 440, '''448''', 450, '''480''', '''512''' * This is the original list for pre-EMAN 2.06: 32, 33, 36, 40, 42, 44, 48, 50, 52, 54, 56, 60, '''64''', 66, 70, '''72''', 81, 84, '''96''', 98, 100, 104, 105, 112, '''120''', '''128''', 130, 132, 140, 150, '''154''', '''168''', 180, 182, 192, '''196''', '''208''', 210, 220, '''224''', '''240''', 250, '''256''', 260, 288, 300, 330, 352, 360, '''384''', 416, 440, '''448''', 450, '''480''', '''512''' * For single particle tomography, the list of "good boxes" is a bit different: 12, 13, 14, 15, 16, 17, 20, 21, 22, 25, 26, 28, 32, 33, 35, 36, 40, 42, 44, 45, 48, 49, 50, 52, 54, 56, 60, 64, 65, 66, 70, 72, 75, 77, 78, 80, 81, 84, 88, 91, 96, 98, 100 Note that if a number is on the list, then 2x the number also tends to be on the list. Since you often use 'shrink=2' when processing. It's a good idea to pick a value twice one of the numbers on the above list. These sizes are less well tested, but also probably good: 540, 576, 600, 625, 640, 648, 675, 720, 729, 750, 768, 800, 810, 864, 900, 960, 972, 1000, 1024, 1080, 1125, 1152, 1200, 1215, 1250, 1280, 1296, 1350, 1440, 1458, 1500, 1536, 1600, 1620, 1728, 1800, 1875, 1920, 1944, 2000, 2025, 2048, 2160, 2187, 2250, 2304, 2400, 2430, 2500, 2560, 2592, 2700, 2880, 2916, 3000, 3072, 3125, 3200, 3240, 3375, 3456, 3600, 3645, 3750, 3840, 3888, 4000, 4050, 4320, 4374, 4500, 4608, 4800, 4860, 5000, 5120, 5184, 5400, 5625, 5760, 5832, 6000, 6075, 6144, 6250, 6400, 6480, 6750, 6912, 7200, 7290, 7500, 7680, 7776, 8000, 8100, ---- |
| Line 3: | Line 28: |
Please also remember that for accurate CTF correction, the box size should be 1.5 - 2x the smallest box that will just contain your particle. Sometimes for large viruses, this requirement is reduced a bit due to speed concerns, but the box size should still be at LEAST 1.25x the size of your particle. |
|
| Line 10: | Line 37: |
| A better way to plot this is with respect to anticipated speed for an N^2 algorithm. This is the reciprocal of the same plot divided by box size squared, | A better way to plot this is with respect to anticipated speed for an O(N^2) algorithm. This is the reciprocal of the same plot divided by box size squared, |
| Line 15: | Line 42: |
Of course, that plot is very difficult to read actual values off of. The original timing data can be downloaded as [[attachment:profile.txt]] From this plot, we can compute when using a larger box-size is better. ie - if you have a box size of 482, your refinement would actually run faster with a box size of 512, even though it's larger. So, when picking a box size, you can optimize your speed by rounding up to a value from this list : 32, 33, 36, 40, 42, 44, 48, 50, 52, 54, 56, 60, 64, 66, 70, 72, 81, 84, 96, 98, 100, 104, 105, 112, 120, 128, 130, 132, 140, 150, 154, 168, 180, 182, 192, 196, 208, 210, 220, 224, 240, 250, 256, 260, 288, 300, 330, 352, 360, 384, 416, 440, 448, 450, 480, 512 Also note that if you are using shrink= it's a good idea to also confirm that your box size divided by the shrink value is in this list. |
Particle Box Size, Reconstruction Quality and Speed
Warning: As a reminder taken from other parts of the documentation: for CTF correction to work well, it is absolutely necessary for the particle box-size to be 1.5-2x the size of the largest axis of your particle. Even if working with stain data, where accurate CTF correction may not be a priority, alignment and other routines requires ~20% padding at a bare minimum. That is, even in these cases a box size 1.2-1.3x the longest particle axis should be considered a minimum. If you go below these values, you may experience a wide range of problems.
Reminder: The appropriate sampling for images for single particle reconstruction is ~2/3 Nyquist. That is, take the best resolution you hope to achieve, and divide by 3. This is close to the optimal A/pix value for your project. If your sampling is worse than this (A/pix larger), then you are not using a high enough magnification on the microscope. If your A/pix is significantly smaller than this (more than 30-50%), you should consider downsampling your data (e2proc2d.py with --meanshrink, for example). For example, if you have 2 A/pix data and are trying to achieve 20 Å resolution you may shrink the data by a factor of 2 or 3. Your refinements will run 5-10x faster, and in some cases, the results will actually be better than those with the finer sampling.
For those who don't like to read (a detailed discussion is below), here is the list of good box sizes: for traditional single particle analysis. Remember that for accurate CTF correction, alignment and reconstruction, the box size should normally be 1.5 - 2x the smallest box that will just contain your particle. Bold numbers also work well with shrinking by 2 or 3:
NEW: This is a list updated on 7/25/2012 for EMAN 2.06 taking into account some recent changes. Note that the new list is shorter. The detailed tables below are still based on the older analaysis.
32, 40, 44, 48, 52, 64, 66, 72, 84, 100, 104, 112, 128, 130, 132, 140, 150, 160, 168, 180, 182, 192, 196, 220, 224, 240, 256, 260, 288, 300, 320, 324, 330, 352, 360, 384, 416, 420, 440, 448, 450, 480, 512
- This is the original list for pre-EMAN 2.06:
32, 33, 36, 40, 42, 44, 48, 50, 52, 54, 56, 60, 64, 66, 70, 72, 81, 84, 96, 98, 100, 104, 105, 112, 120, 128, 130, 132, 140, 150, 154, 168, 180, 182, 192, 196, 208, 210, 220, 224, 240, 250, 256, 260, 288, 300, 330, 352, 360, 384, 416, 440, 448, 450, 480, 512
- For single particle tomography, the list of "good boxes" is a bit different:
12, 13, 14, 15, 16, 17, 20, 21, 22, 25, 26, 28, 32, 33, 35, 36, 40, 42, 44, 45, 48, 49, 50, 52, 54, 56, 60, 64, 65, 66, 70, 72, 75, 77, 78, 80, 81, 84, 88, 91, 96, 98, 100
Note that if a number is on the list, then 2x the number also tends to be on the list. Since you often use 'shrink=2' when processing. It's a good idea to pick a value twice one of the numbers on the above list.
These sizes are less well tested, but also probably good:
540, 576, 600, 625, 640, 648, 675, 720, 729, 750, 768, 800, 810, 864, 900, 960, 972, 1000, 1024, 1080, 1125, 1152, 1200, 1215, 1250, 1280, 1296, 1350, 1440, 1458, 1500, 1536, 1600, 1620, 1728, 1800, 1875, 1920, 1944, 2000, 2025, 2048, 2160, 2187, 2250, 2304, 2400, 2430, 2500, 2560, 2592, 2700, 2880, 2916, 3000, 3072, 3125, 3200, 3240, 3375, 3456, 3600, 3645, 3750, 3840, 3888, 4000, 4050, 4320, 4374, 4500, 4608, 4800, 4860, 5000, 5120, 5184, 5400, 5625, 5760, 5832, 6000, 6075, 6144, 6250, 6400, 6480, 6750, 6912, 7200, 7290, 7500, 7680, 7776, 8000, 8100,
Various algorithms in EMAN2 will depend non-linearly on the box size of the particle. Sometimes (such as the case with FFTs), this behavior will appear bizzare. For example refinements with a box size of 45 pixels will run roughly twice as fast as those with a box size of 47, and 44 is about 20% faster than 45.
Please also remember that for accurate CTF correction, the box size should be 1.5 - 2x the smallest box that will just contain your particle. Sometimes for large viruses, this requirement is reduced a bit due to speed concerns, but the box size should still be at LEAST 1.25x the size of your particle.
The following plot shows how long it takes to compute one similarity matrix element for a noisy particle aligned to a noise-free reference with the rotate-translate-flip aligner, refine alignment enabled with the dot comparator, and a phase residual for a similarity metric. ie - typical options for a real refinement:
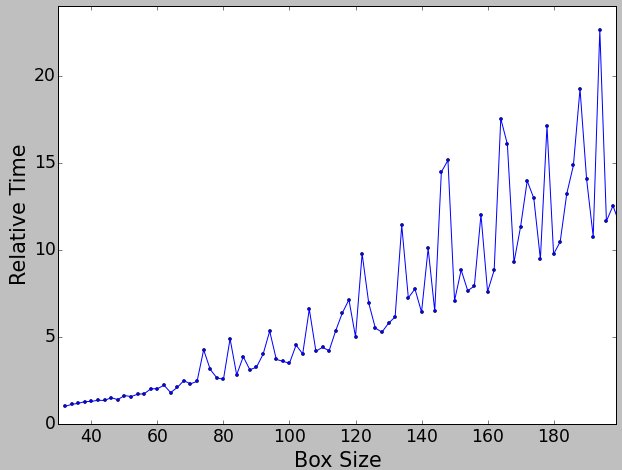
Clearly there are some good box sizes, and some very bad box sizes.
A better way to plot this is with respect to anticipated speed for an O(N^2) algorithm. This is the reciprocal of the same plot divided by box size squared, normalized so 512 is 1. That is, larger values indicate better relative speeds. Of course, 103 is still faster than 512, but if you look in a local neighborhood for a peak, that will correspond to a good box size to use.
Of course, that plot is very difficult to read actual values off of. The original timing data can be downloaded as profile.txt
From this plot, we can compute when using a larger box-size is better. ie - if you have a box size of 482, your refinement would actually run faster with a box size of 512, even though it's larger. So, when picking a box size, you can optimize your speed by rounding up to a value from this list :
32, 33, 36, 40, 42, 44, 48, 50, 52, 54, 56, 60, 64, 66, 70, 72, 81, 84, 96, 98, 100, 104, 105, 112, 120, 128, 130, 132, 140, 150, 154, 168, 180, 182, 192, 196, 208, 210, 220, 224, 240, 250, 256, 260, 288, 300, 330, 352, 360, 384, 416, 440, 448, 450, 480, 512
Also note that if you are using shrink= it's a good idea to also confirm that your box size divided by the shrink value is in this list.
