|
Size: 182
Comment:
|
Size: 622
Comment:
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 1: | Line 1: |
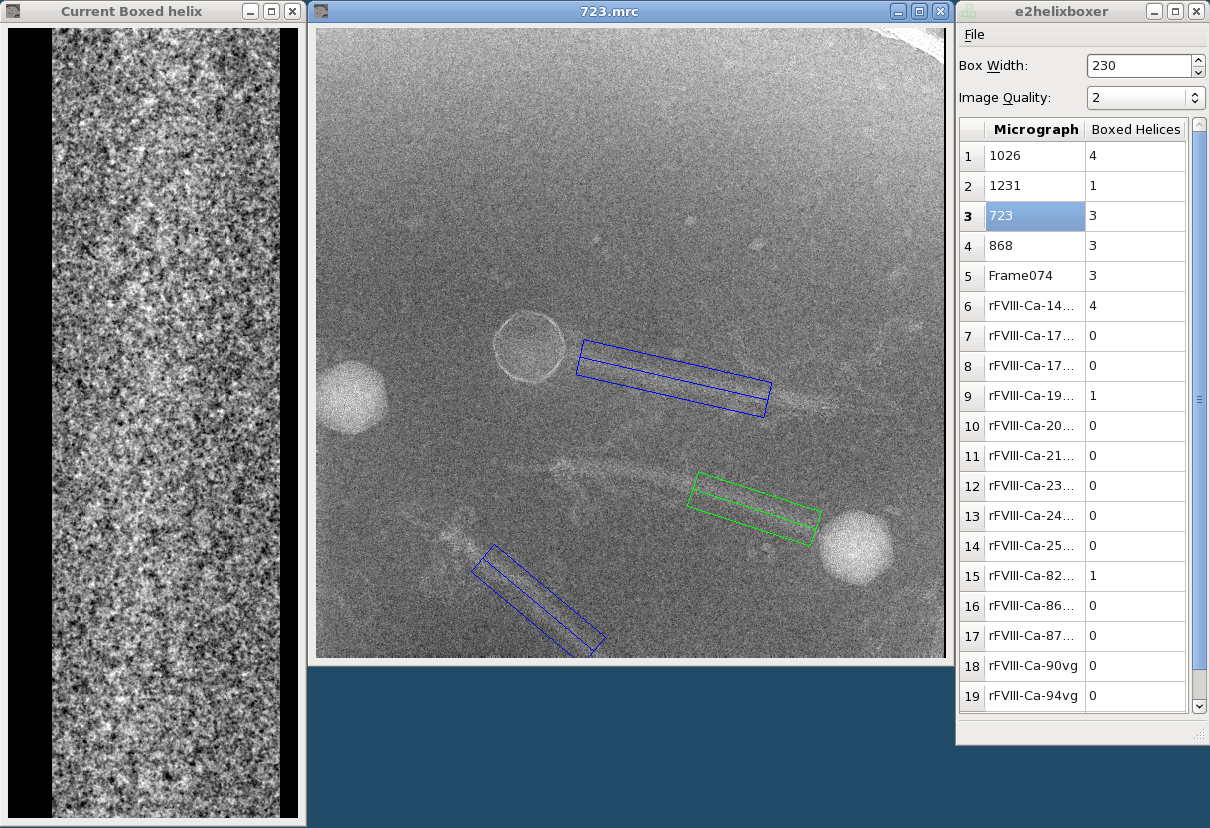
| e2helixboxer.py is used to select rectangular 2D projections of helices from a micrograph, and extract overlapping particles from the boxed helices. {{attachment:e2helixboxer.png}} | = e2helixboxer.py: Overview = e2helixboxer.py is used to select rectangular 2D projections of helices from a micrograph, and extract overlapping particles from the boxed helices. == GUI mode == To start the program's graphic user interface, use the "--gui" option. You can follow this with a micrograph filename, a list of micrograph filenames, or nothing. {{{ $ e2helixboxer.py --gui <micrograph1> <micrograph2> <...> }}} {{{ $ e2helixboxer.py --gui $ e2helixboxer.py --gui 101.mrc $ e2helixboxer.py --gui *.mrc micrograph.hdf *.img abc.dm3 }}} {{attachment:e2helixboxer.png}} == Command-line mode == |
e2helixboxer.py: Overview
e2helixboxer.py is used to select rectangular 2D projections of helices from a micrograph, and extract overlapping particles from the boxed helices.
GUI mode
To start the program's graphic user interface, use the "--gui" option. You can follow this with a micrograph filename, a list of micrograph filenames, or nothing.
$ e2helixboxer.py --gui <micrograph1> <micrograph2> <...>
$ e2helixboxer.py --gui $ e2helixboxer.py --gui 101.mrc $ e2helixboxer.py --gui *.mrc micrograph.hdf *.img abc.dm3