|
Size: 175
Comment:
|
Size: 1188
Comment:
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 6: | Line 6: |
| Programs in the tomogram segmentation requires Theano, which is not distributed with EMAN2. | Programs in the tomogram segmentation requires Theano, which is not distributed with EMAN2. To use the protocol, one needs to build EMAN2 from source and install Theano manually. http://deeplearning.net/software/theano/install.html == Getting Started == First, make an empty directory and get into that directory in command line. Then run e2projectmanager.py from the command line. While a GUI window will show up, it is still a good idea to keep the command line window open to view the messages. Click the '''Workflow Mode''' drop-down menu next to navigate to the TomoSeg panel. {{attachment:tomoseg_panel.png}} == Import Tomograms == == Select Positive Samples == == Manually Annotate Samples == == Select Negative Samples == == Build Training Set == == Train Neural Network == == Apply to Tomograms == == Acknowledgement == Darius Jonasch, the first user of the tomogram segmentation protocol, provided many useful advices to make the workflow user-friendly. He also wrote a tutorial of the earlier version of the protocol, on which this tutorial is based. |
Tomogram Segmentation
Availability: EMAN2 daily build after 2016-10
Programs in the tomogram segmentation requires Theano, which is not distributed with EMAN2. To use the protocol, one needs to build EMAN2 from source and install Theano manually.
http://deeplearning.net/software/theano/install.html
Getting Started
First, make an empty directory and get into that directory in command line. Then run e2projectmanager.py from the command line. While a GUI window will show up, it is still a good idea to keep the command line window open to view the messages.
Click the Workflow Mode drop-down menu next to navigate to the TomoSeg panel. 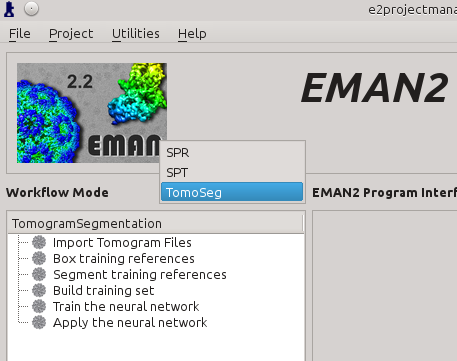
Import Tomograms
Select Positive Samples
Manually Annotate Samples
Select Negative Samples
Build Training Set
Train Neural Network
Apply to Tomograms
Acknowledgement
Darius Jonasch, the first user of the tomogram segmentation protocol, provided many useful advices to make the workflow user-friendly. He also wrote a tutorial of the earlier version of the protocol, on which this tutorial is based.
