|
Size: 18738
Comment: update to protocol exchange
|
Size: 18856
Comment: add rutgers tutorial
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 5: | Line 5: |
| If you are using this method in a publication, please cite: | If you are using this in a publication, please cite: |
| Line 11: | Line 11: |
| * 01/2018: Slides of a recent tutorial in Rutgers boot camp can be found here. [[attachment:tomoseg_tutorial_rutgers.pdf]] |
Tomogram Annotation/Segmentation
Availability: EMAN2.2+, or daily build after 2016-10 This is the tutorial for a Convolutional neural network(CNN) based semi-automated cellular tomogram annotation protocol.
If you are using this in a publication, please cite:
M. Chen et al. (2017). Convolutional Neural Networks for Automated Annotation of Cellular Cryo-Electron Tomograms. Nature Methods, http://dx.doi.org/10.1038/nmeth.4405
A stable (thus not up-to-date) online version of the tutorial can also be found at Protocol exchange: https://www.nature.com/protocolexchange/protocols/6171
Updates:
01/2018: Slides of a recent tutorial in Rutgers boot camp can be found here. tomoseg_tutorial_rutgers.pdf
- 10/2017: We have switched to gpuarray backend for CUDA9 support. It should still work with CUDA7.5+ and the old cuda backend in Theano.
Installation
The CNN based tomogram annotation protocol works best on a workstation with a CUDA compatible GPU. While the software is still able to function on a single core of a CPU at a gratly reduced speed, the time required makes this impractical. The GPU needs to be equipped with sufficient memory to store the training set as well as the CNN. This is roughly 1 GB with default parameters but could be larger in some situations. Ideally, a dedicated GPU should be used for computation with a second, generally lower end, GPU for display.
Given the effective requirement of CUDA, at present, Linux is the only recommended platform, though emerging support for OpenCL means that accelerated support for the Mac may be possible in the near future.
Since the setup of GPU computing environment closely depends on the hardware and operating system, this is an independent process from installation of EMAN2. Instructions for installing current versions of CUDA change frequently, but can be found on the Nvidia website at https://developer.nvidia.com/cuda-toolkit. For some Linux distributions, CUDA can also be simply installed via built in package manager.
The CNN implementation in this software is based on the Python library Theano. Although Theano is already installed during EMAN2 installation, it still needs to be configured to use the correct GPU environment after CUDA setup. A guide for GPU usage in Theano can be found at http://deeplearning.net/software/theano/tutorial/using_gpu.html.
Some extra information on how to use CUDA and GPU can be found at http://blake.bcm.edu/emanwiki/EMAN2/Install/BinaryInstallAnaconda#Using_the_GPU
Setup workspace
First, make an empty folder and cd into that folder at the command-line. Run e2projectmanager.py. While a GUI window will appear, messages may appear in the terminal used to launch the program, so it is useful to keep this window visible.
Click the Workflow Mode drop-down menu next to navigate to the TomoSeg panel.
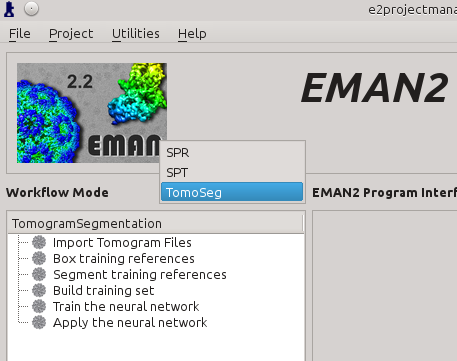
Import Tomograms
Click Import Tomogram Files on the left panel (1). On the panel showed up on the right, click Browse next to import_files (2), and select the tomogram you would like to segment in the browser window, and click Ok. If you want to downsample/bin the tomogram before processing, write the shrinking factor in appropriate box (3). Also make sure that the import_tomos and tomoseg_auto box is checked. Finally, click Launch (4) and wait the pre-process to finish.
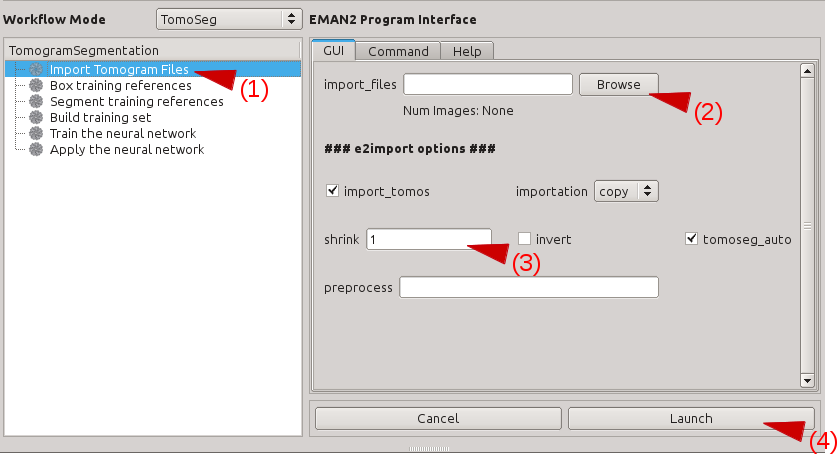
- This process pre-filter and normalize the imported tomograms, so it is critical to import all tomograms you need to process in the same way, including the tomogram used for picking patches and training, as well as other tomograms you wish to apply the trained CNN on. Performance of the CNN can be unpredictable if the tomograms are not filtered and normalized properly.
- The CNN should be able to recognize features that are larger than 2 pixels, and downsampling the tomogram by 2 will speed the annotation process by 8. So you can downsample the tomogram heavily as long as you can still recognize the features by eye.
- When you want to annotate features of multiple scales, for example actin filaments and viruses in a tomogram, if you downsample by the same factor, either the small features will be invisible, or the large features cannot fit in the 64x64 patch. In these cases, you can import the tomogram twice with different downsample factor, annotate features in the imported tomograms separately, up-scale the annotation results of the smaller tomogram and merge the annotations.
Select Positive Samples
Select Box training references. Press browse, and select an imported tomogram. Leave boxsize as -1, and press Launch.
In a moment, three windows will appear: the tomogram view, the selected particles view, and a control-panel. Unlike the 2-D particle picking interface used in single particle analysis, which is almost identical, this program allows you to move through the various slices of the tomogram. The examples you select will be 2-D, drawn from arbitrary slices throughout the 3-D tomogram.
In the window named e2boxer, make sure your box size is 64. None of the other options need to be changed.
In the window containing your tomogram, you can begin selecting boxes. You can move through the slices of the tomogram with the up and down arrows, and zoom in and out with the scroll wheel. Alternatively, you can middle-click on the image to open a control-panel containing sliders with similar functionality. Select and reposition regions of interest (ROIs) with the left mouse button. Hold down shift when clicking to delete existing ROIs. As you select regions, they will appear in the (Particles) window.
Select around 10 ROIs containing the feature you wish to annotate. You will repeat this training process from scratch for different features, so for now, focus just on a single feature type. If the same feature appears different in different layers or different regions of the cell, be sure to cover the full range of visual differences in the representative regions you select. When selecting boxes, ensure that feature is clear in the (Particles) window. You will manually identify the feature in the next step, so it is critical that you can accurately identify the feature throughout each ROI. It is better to have fewer ROIs that you can annotate well than more ROIs with ambiguous annotations.
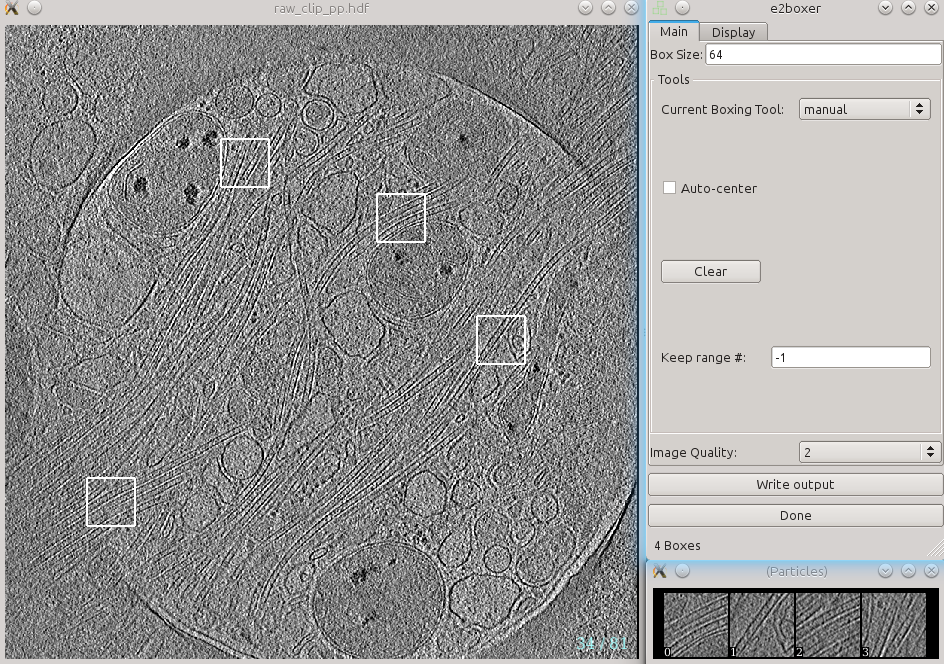
After identifying an appropriate number of boxes, press Write output in the e2boxer window.
Select your tomograms in the Raw Data window.
Select a suffix for the ROIs in the Output Suffix text box (perhaps _good).
In Normalize Images, select None.
Press OK.
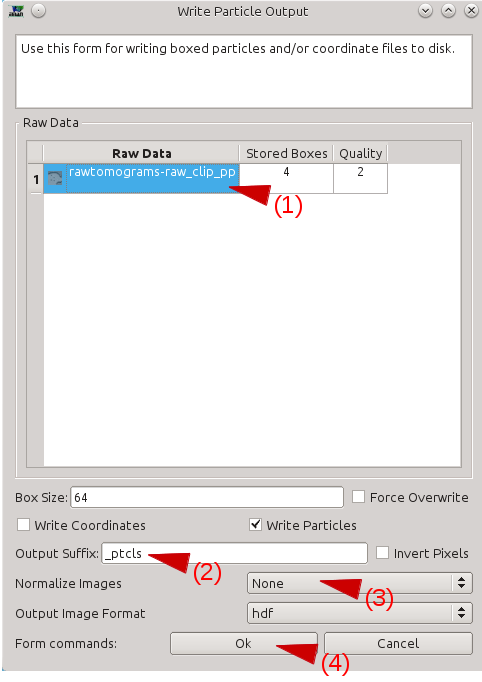
- Make sure the box size is 64. If the feature of interest does not fit in a 64x64 tile, try downsample your tomogram more.
Manually Annotate Samples
The next step is to manually identify the feature within each ROI.
Navigate to the Segment training references interface in the project manager window. For Particles, browse to the output ROIs you generated in the previous step. Leave Output blank, keep segment checked, and press Launch.
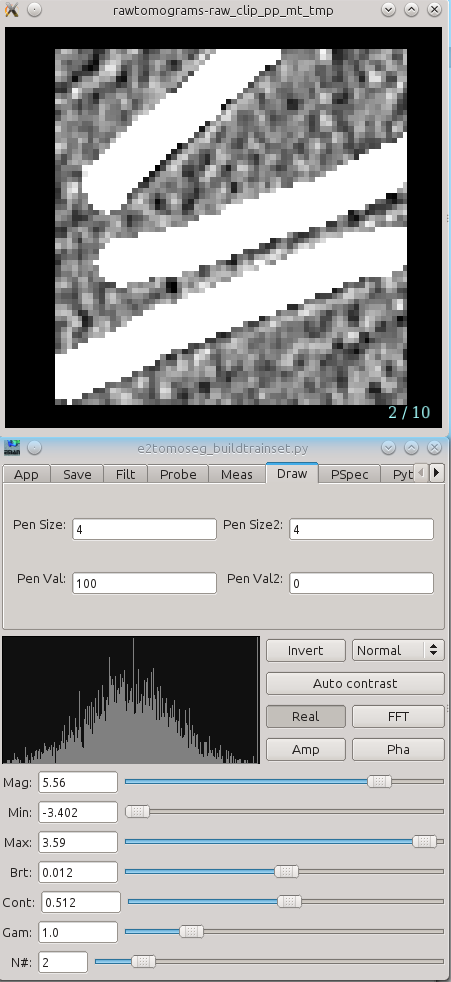
Two windows will appear, one containing images, and the other a control panel. You can navigate through the images similarly to looking through the slices of the tomogram above. The control panel will open with the Draw tab selected. Using the left mouse button, draw on each of the ROIs to exactly cover the feature of interest as best you can. If necessary you can always can go back to the ROI selection window and check the surroundings of each region to aid in segmentation.
Segment all of your ROIs. If there are multiple ROIs in one 2D sample, like multiple microtubules in this case, make sure to segment all of them. If you need to change the size of the pen, change both Pen Size and Pen Size2 to a larger or smaller number. You would like the marked region to match the feature as closely as possible, so reduce the pen size if it is larger than the feature.
When you are finished, simply close the windows. The segmentation file will be saved automatically as "*_seg.hdf" with the same base file name as your ROIs.
- Generally speaking, the CNN can recognize the features if you can recognize the same feature in 2D patches by eye without ambiguity. Note that you need to be able to recognize features in 2D patches locally, i.e. if in one piece of micrograph you see a fragment of microtubule on the left, a fragment of microtubule on the right but it is too noisy to see anything in the center, the CNN is unlikely to work if you assume there are microtubule in the center and use that as training set.
- If you are not certain about your manual annotation in one sample, just do not pick that 2D patch for training. Only train the network on features that you are certain that are positive.
- Do not leave positive features un-annotated. If you are annotating ribosomes and there are 5 of them in one sample, make sure to segment all 5 of them. Since we only require a small amount of positive training set, each one of them is critical.
Select Negative Samples
Next you need to identify regions in the tomogram which do not contain the feature of interest at all. Return to the same interface you used to select Positive examples, and press the Clear button in the e2boxer window. This deletes all of your previous selections (the positive examples), so make sure you have finished the previous steps before doing this.
In the tomogram window, select boxes that DON’T contain the feature of interest. You can select as many of these as you like (normally ~100). Try to include a wide variety of different non-feature cellular structures, empty space, gold fiducials and high-contrast carbon.
After finishing picking the negative samples, write the particle output in same way you did for the positive samples, but use a different Output Suffix, like "_bad".
Build Training Set
Select the Build training set option in EMAN2.
In particles_raw, select your "_ptcls" file.
In particles_label, select your "_ptcls_seg" file.
In boxes_negative, select your "_bad" file.
Leave trainset_output blank. Ncopy controls the number of particles in your training set. The default of 10 is fine, unless you want to do a faster run at the expense of accuracy.
Press Launch. The program will print "Done" in your Terminal when it has finished. The training set will be saved as the same name as the positive particles with "_trainset" suffix.
Note that if you have not configured Theano to use CUDA as described above, this will run on a single CPU and take a VERY long time.
Train Neural Network
Open Train the neural network in the project manager. In trainset, browse and choose the "_trainset" file.
The defaults for everything else in this window are sufficient to produce good results. To significantly shorten the length of the training (but potentially reduce the quality), reduce the number of iterations. Write the filename of the trained neural network output in the netout text box, and leave the from_trained box empty if it is the first training process.
Press Launch. The program will print a few numbers quickly at the beginning (this is to monitor the training process. Something is wrong if it prints really huge values), and then will notify you once it's completed each iteration. When it's finished, it will output the trained neural network in the specified netout file and samples of the training result in a file called "trainout_" followed by the netout file name.
After the training is finished, it is recommended to look at the training result before proceeding. Open the "trainout_*.hdf" file from the e2display window (use show stack), and you should see something like this.
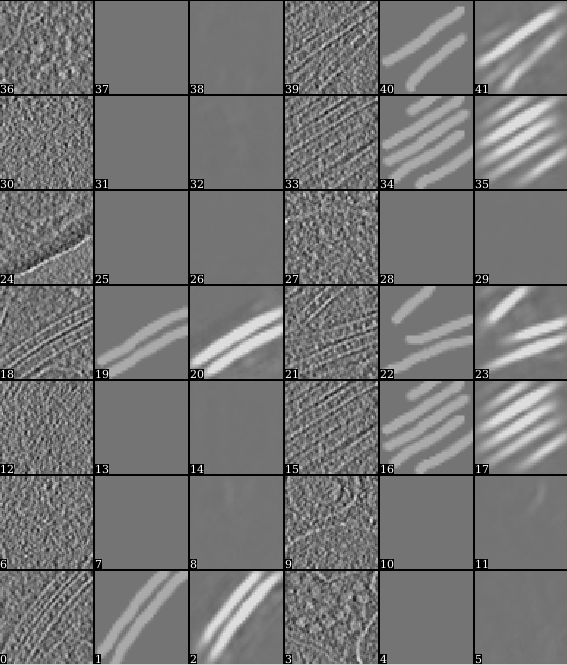
Zoom in or out a bit so there are 3*N images displayed in each row. For each three images, the first one is the ROI from the tomogram, the second is the corresponding manual segmentation, and the third is the neural network output for the same ROI after training. The neural network is considered well trained if the third image matches the second image. For the negative particles, both the second and the third images should be largely blank (the 3rd may contain some very weak features, this is ok).
If the training result looks somewhat wrong, go back and check your positive and negative training set first. Most significant errors are caused by training set errors, i.e. having some positive particles in the negative training set, or one of the positive training set is not correctly segmented. If the training result looks suboptimal (the segmentation is not clear enough but not totally wrong), you may consider continuing the training for a few rounds. To do this, go back to the Train the neural network panel, choose the previously trained network for the from_trained box and launch the training again. It is usually better to set a smaller learning rate in the continued training. Consider change value in the learnrate to ~0.001, or the displayed learning rate value at the last iteration of the previous training.
If you are satisfied with the result, go to the next step to segment the whole tomogram.
Apply to Tomograms
Finally, open Apply the neural network panel. Choose the tomogram you used to generate the boxes in the tomograms box, choose the saved neural network file (not the "trainout_" file, which is only used for visualization), and set the output filename. You can change the number of threads to use by adjusting the thread option. Keep in mind that using more threads will consume more memory as the tomogram slices are read in at the same time. For example, processing a 1k x 1k x 512 downsampled tomogram on 10 cores would use ~5 GB of RAM. Processing an unscaled 4k x 4k x 1k tomogram would increase RAM usage to ~24 GB. When this process finishes, you can open the output file in your favourite visualization software to view the segmentation.
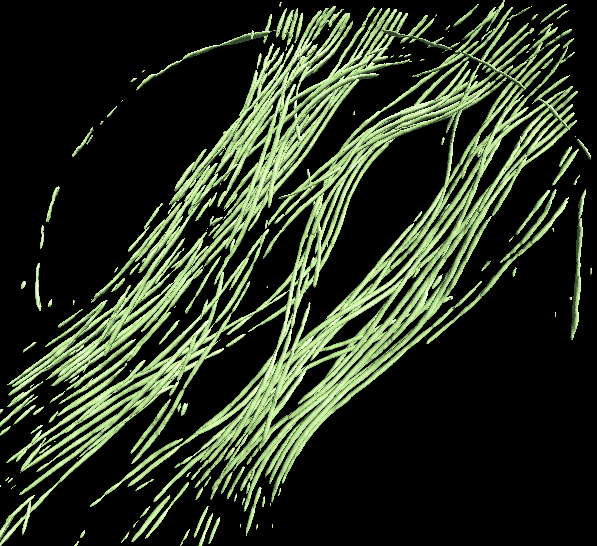
To segment a different feature, just repeat the entire process for the each feature of interest. Make sure to use different file names (eg - _good2 and _bad2)! The trained network should generally work well on other tomograms using a similar specimen with similar microscope settings (clearly the A/pix value must be the same).
Merging multiple annotation results
Merging the results from multiple networks on a single tomogram can help resolve ambiguities, or correct regions which were apparently misassigned. For example, in the microtubule annotation shown above, the carbon edge is falsely recognized as a microtubule. An extra neural network can be trained to specifically recognize the carbon edge and its result can be competed against the microtubule annotation. A multi-level mask is produced after merging multiple annotation result in which the integer values in a voxel identify the type of feature the voxel contains. To merge multiple annotation results, simply run in the terminal:
mergetomoseg.py <annotation #1> <annotation #2> <...> --output <output mask file>
Tips in selecting training samples
- Annotate samples correctly, as a bad segmentation in the training set can damage the overall performance. In the microtubule case, if you annotate the spacing between microtubules, instead of the microtubules themselves (it is actually quite easy to make such mistake when annotating microtubule bundles), the neural network can behave unpredictably and sometimes just refuse to segment anything. Here is the training result on an incorrect and correct segmentation in one training set. Note the top one (22) is annotating the space between microtubules.

- Make sure there are no positive samples in the negative training set. If your target feature is everywhere and it is hard to find negative regions, you can add additional positive samples which include various features other than the target feature (annotating only the target feature).
- You can bin your tomogram differently to segment different features. Just import multiple copies of raw tomogram with different shrink factors, and unbin the segmentation using math.fft.resample processor. It is particularly useful when you have features with different lengthscales in one tomogram, and it is impossible to both fit the large features into a 64*64 box and still have the smaller features visible at the same scale.
- In some cases, there is a significant missing wedge in the x-y plane slices (you can visualize this by clicking Amp button when looking at the slices in EMAN2). So the resolvability on x direction is different than that on y direction. It is important to provide features running in different directions in the training set, otherwise the neural net may only pick up features in one direction based on the Fourier patten. Also, you may want to check the stage of the microscope, since this may suggest the sample is not tilted exactly around the x axis.
- It is also vital to cover various states of the target feature. For example, if you want to segment single layer membranes, you may want to have some cell membrane, some small vesicles, and some vesicles with darker density inside, so the neural network can grab the concept of membrane. Just imagine how you would teach someone with no biological knowledge about the features you are looking for. On the other hand, it is possible to ask the neural network to separate different types of those same features. In the membrane example, it is possible to train the neural network to segment vesicles from cell membranes based on the curvature, or recognize dense vesicles based on the difference of intensity on both side of the membrane, given carefully picked training set.
Acknowledgement
Darius Jonasch, the first user of the tomogram segmentation protocol, provided many useful advices to make the workflow user-friendly. He also wrote a tutorial of the earlier version of the protocol, on which this tutorial is based.
