Tomogram Segmentation
Availability: EMAN2 daily build after 2016-10
Programs in the tomogram segmentation requires Theano, which is not distributed with EMAN2. To use the protocol, one needs to build EMAN2 from source and install Theano manually.
http://deeplearning.net/software/theano/install.html
Getting Started
First, make an empty directory and get into that directory in command line. Then run e2projectmanager.py from the command line. While a GUI window will show up, it is still a good idea to keep the command line window open to view the messages.
Click the Workflow Mode drop-down menu next to navigate to the TomoSeg panel.
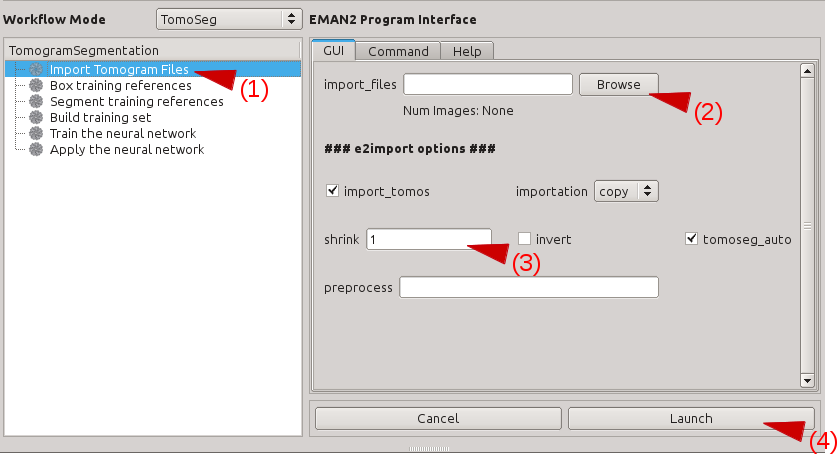
Import Tomograms
Click "Import Tomogram Files" on the left panel. On the panel showed up on the right, click "Browse" next to import_files, and select the tomogram you would like to segment in the browser window, and click "Ok". If you want to bin the tomogram before processing, write the shrinking factor in the text box next to "shrink". Make sure that the "import_tomos" and "tomoseg_auto" box is checked. Finally, click "Launch" and wait the pre-process to finish.
Select Positive Samples
Open "Box training references". Press browse, and select your imported tomogram. Leave “boxsize” at -1, and press Launch.
In a moment, three windows will appear on your screen, which will be familiar if you’ve boxed particles before. The only difference between this boxing and the other is that you can box in 2D on slices of a 3D image.
On the window named “e2boxer”, make sure your box size is 64. None of the other options need to be changed.
On the window containing your tomogram, you can begin selecting boxes. Go up and down in the tomogram using the arrow keys, select and drag boxes using the left mouse button, and delete boxes using Shift + left mouse button. As you select boxes, they will appear in the (Particles) window.
Select around 10 boxes containing your structure. If your structure appears differently throughout the cell (e.g. microtubules), be sure to include a variety of views in the boxes.
When selecting boxes, ensure that your structure is clear in the (Particles) window. You will have to manually segment these boxes, so if you can’t see your structure, your segmentation will be more difficult, and your final segmentation will suffer as a result. It is better to have fewer boxes that you can segment better than more boxes you segment worse.
After getting an appropriate number of boxes, press “Write output” in the e2boxer window.
- 1 Select your boxes in the “Raw Data” window. 2 Change the “Box Size” to the box size you want. 3 Select the “Force Overwrite” box. 4 Leave the “Output Suffix” as _ptcls. 5 In “Normalize Images”, select “None”. 6 Press “OK”.
Manually Annotate Samples
Select Negative Samples
Build Training Set
Train Neural Network
Apply to Tomograms
Acknowledgement
Darius Jonasch, the first user of the tomogram segmentation protocol, provided many useful advices to make the workflow user-friendly. He also wrote a tutorial of the earlier version of the protocol, on which this tutorial is based.
