|
Size: 8493
Comment: focuesed refinement
|
Size: 8402
Comment:
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 58: | Line 58: |
| * This will give us a slightly better alignment on the head part of the ribosome. Once the refinement finishes, we can run a subsequent subtilt refinement based on the subtomogram refinement. There is nothing really different here so just start a run from the GUI. We should get something like this, better than the head part in the original refinement, yet still not quite as good as the large subunit of the ribosome. {{attachment:ribo_small_refine.png| Refined ribosome head part |width=600}} |
* This will give us a slightly better alignment on the head part of the ribosome. Once the refinement finishes, we can run a subsequent subtilt refinement based on the subtomogram refinement. There is nothing really different here so just start a run from the GUI. We should get something better than the head part in the original refinement, yet still not quite as good as the large subunit of the ribosome. |
Extra functions for EMAN2 tomography
- This page describes extra functionalities of EMAN2 tomography workflow. This tutorial is frequently updated, so it is better to have EMAN2 version as new as possible.
Focused refinement
Refine local regions of a large complex. Available after 05/23/2019. Still under development.
Start from a successful subtomogram refinement of the full complex (spt_xx folders). For large complexes, it is more convenient to run the full complex refinement with bin2 or smaller particles, then run local refinement with unbinned particles. Here we start from the result from the ribosome tutorial dataset: e2tomo.
Here we follow the Relion single particle multibody refinement paper and focus on the "head" part of the ribosome. From the current subtomogram averaging result, it is quite obvious that the head part has a worse resolution. The great thing about tomography here is that, since we have 3D information of each individual particle, we can just mask out the region we like, instead of doing particle subtraction.
First we need to locate the region of interest. Here we use the filter tool to do it interactively. Run Fitertool on the 3D structure we currently have and insert a mask.soft processor. Play with the parameters. Here I have dx=32:dy=32:dz=0 and the radius of the head part is roughly 36 pixels.
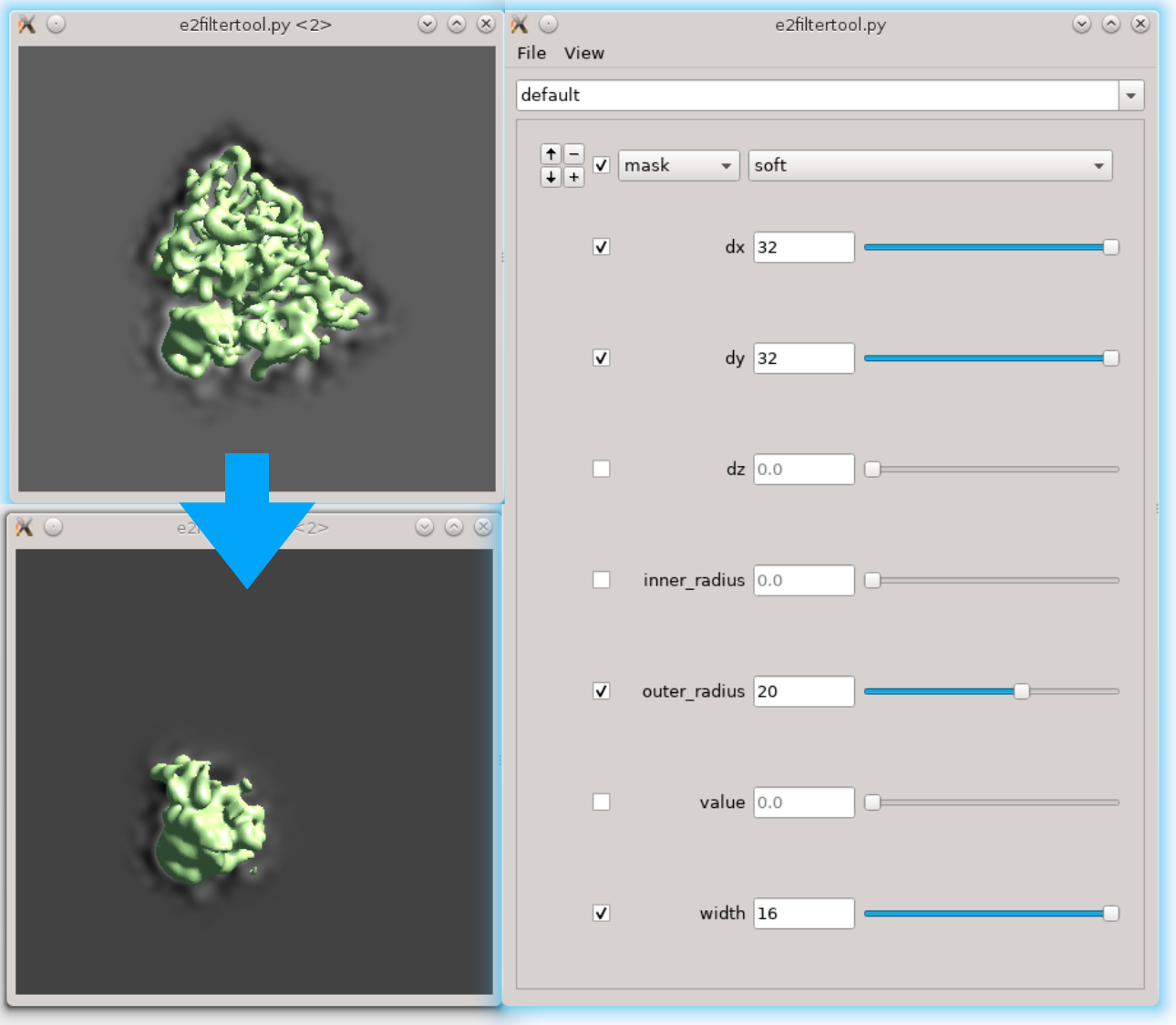
- Now clip the region out and take a look. The box size of the local region needs to be at least 72, but we use 128 regardless because extra padding is always good. Assume we have the result in spt_01 and iteration 5. Run
e2proc3d.py spt_01/threed_05.hdf ref_small_0.hdf --clip 128,128,128,128,128,96
Here the first three numbers after --clip are the box size in x,y,z, and the next three numbers are the center of the box. My original box size is 192, so the center is at 96,96,96. Since we decided that the shift is 32,32,0 in the previous step, the new center should be at 128,128,96.
Next we need to make a mask of the region. Open Filtertool on the map we just generated (ref_small_0.hdf), and make a mask you like. Try making the mask as soft as possible but do not touch the edge of the box. Here I use a mask.soft with mask.auto3d.thresh. Once you are satisfied, save the map from Filtertool in File -> Save processed Map. The map will be saved as processed_map.hdf, and we rename it to mask_small_0.hdf. Make sure we save the mask file (with values between 0 and 1), not the masked map. We can also make a masked reference by multiplying the reference with the mask.
e2proc3d.py ref_small_0.hdf ref_small_0_mask.hdf --multfile mask_small_0.hdf

- We also need to prepare the transform input for particle extraction. Open a new text file and write
{'type':'eman', 'tx':32, 'ty':32, 'tz':0}Save the file as xf_small.txt. This seems overcomplicated because this framework also allows local refinement of multiple asymmetrical units individually. For details on how to do this, check the next section.
- Now run particle extraction to get those sub-particles. Run
e2spt_extract.py --jsonali spt_01/particle_parms_05.json --boxsz_unbin 160 --newlabel ribo_small --postxf xf_small.txt --postmask mask_small_0.hdf
Here the program will go back to the original tilt series, extract 2D subtilts and reconstruct the 3D particles. So you can extract unbinned particles even if your initial refinement is done on downsampled particles. The program will deal with scaling automatically. CTF information will also be calculated again, with per-particle defocus at the center of the sub-particles used. The program will not actually align the particles, it will only save the alignment parameters in the header as xform.align3d.
Make a particle set from the newly extracted particles. Here we will have sets/ribo_small.lst. To make sure we are doing everything correctly, average the particles together using the transform in their header.
e2proc3d.py sets/ribo_small.lst avg_ribo_small.hdf --average --averager mean.tomo --avgbyxf
Hopefully, the average should look like the reference we make from the previous subtomogram averaging earlier (you may need to lowpass filter it a bit). If there are many particles, e2proc3d.py can be quite slow as it is single threaded. There is a program in the examples folder that can parallelize the process. If you have examples in your path, run
e2procnd_par.py "e2proc3d.py sets/ribo_small.lst avg_ribo_small.hdf --average --averager mean.tomo --avgbyxf" --threads 24
This also works on most other e2proc2d and e2proc3d processes.
- If everything is working, we can start the refinement run with the particles and the reference.
e2spt_refine.py sets/ribo_small.lst --reference ref_small_0_mask.hdf --niter 5 --maxalt 45 --refine
You can use any e2spt_refine.py options you like, just make sure to add the --refine option. When the mask used for the particle extraction step is not soft enough, it might affect the missing wedge determination, so setting a --maxalt seems to be a safer choice.
- This will give us a slightly better alignment on the head part of the ribosome. Once the refinement finishes, we can run a subsequent subtilt refinement based on the subtomogram refinement. There is nothing really different here so just start a run from the GUI. We should get something better than the head part in the original refinement, yet still not quite as good as the large subunit of the ribosome.
- We can also look at the alignment difference in the focused refinement comparing to the initial full ribosome refinement. An eigen-motion trajectory can be built from the refinement to show how the subunit moves with respect to other parts of the ribosome. Run
e2spt_trajfromrefine.py --path spt_02 --iter 5
We can visualize the motion in Chimera.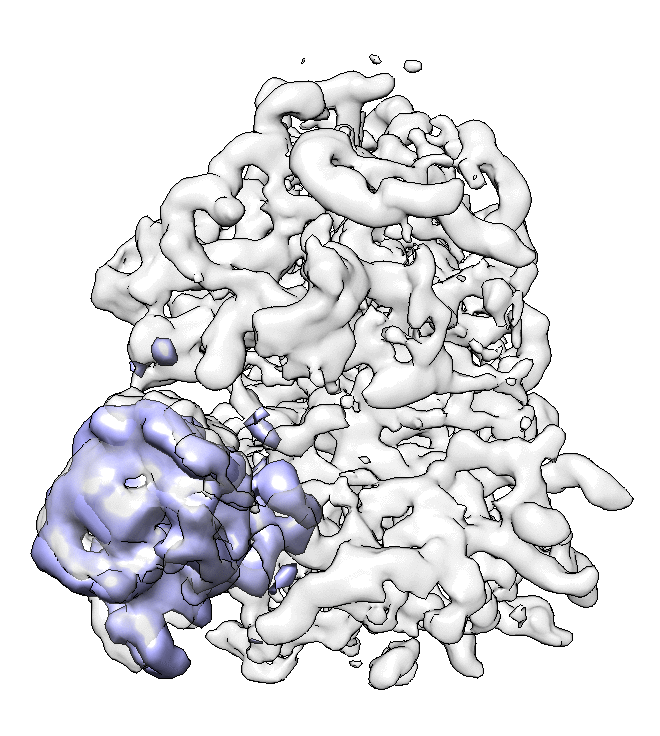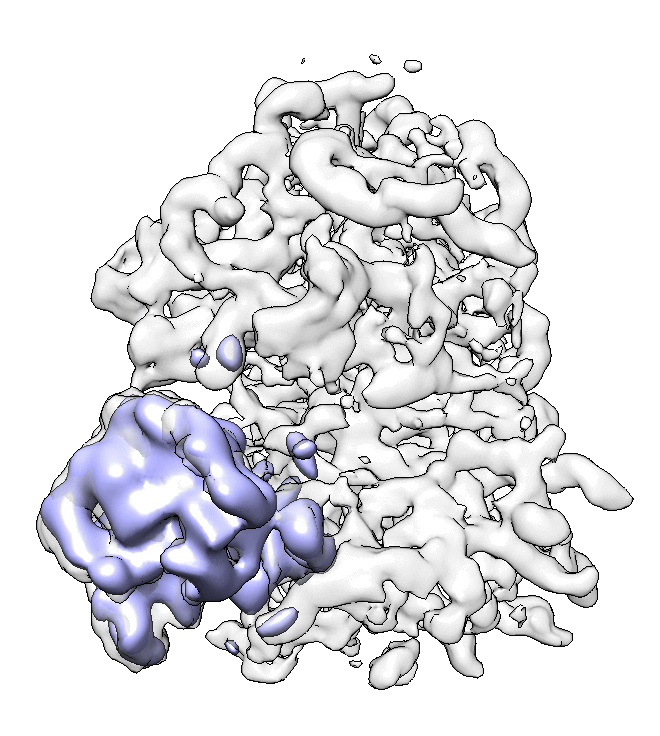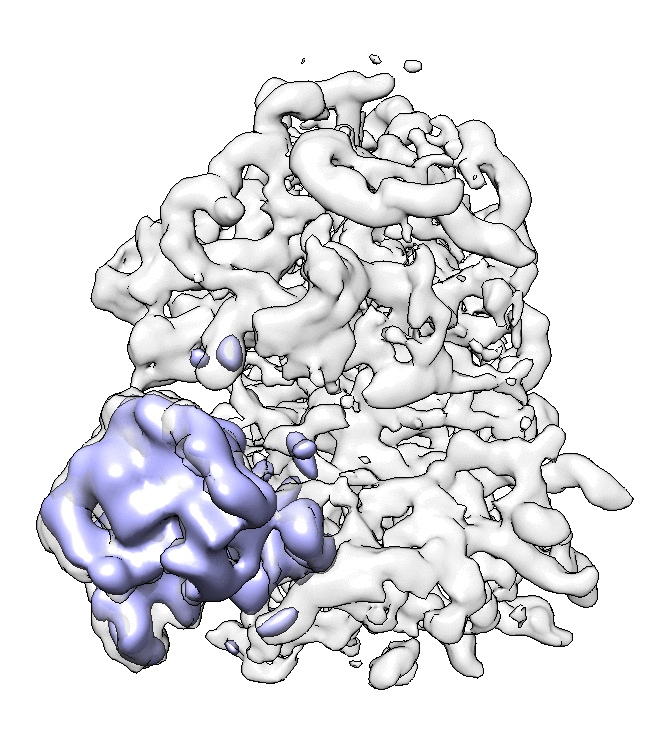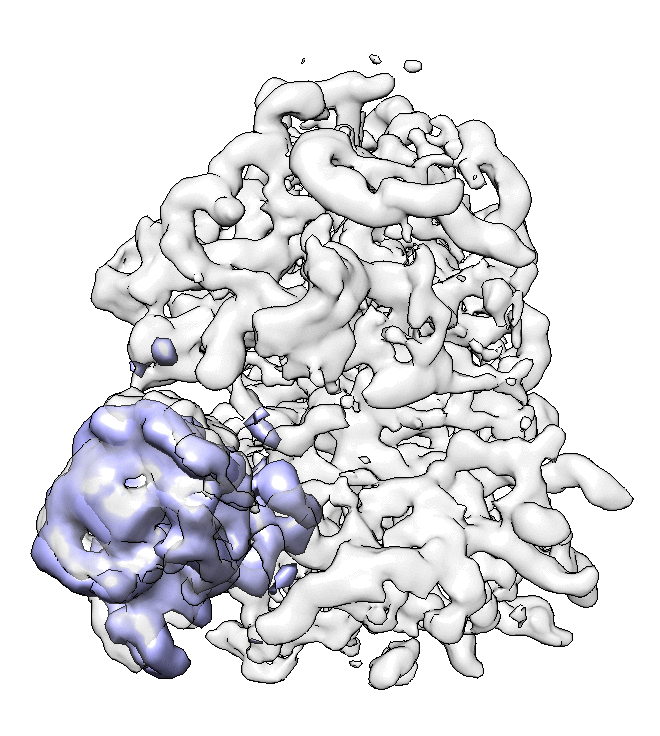
Focused refinement on multiple asymmetrical units
When you have a complex with multiple asymmetrical units, start from one unit and get the transform and mask following the previous section. Assume we have c5 symmetry and the first unit is at 32,32,0. Then run e2.py and type
sym=Symmetries.get("c5") xf0=Transform({"type":"eman", "tx":32, "ty":32, "tz":0}) for i in range(sym.get_nsym()): xf=sym.get_sym(i) print((xf0*xf).get_params("eman"))
You will get a list of transform dictionaries in the printout. Paste them into a text file and use it as the input for particle extraction.
This also works when you have a complex with multiple identical components but does not follow a clear symmetry. Extract each unit individually and align the same reference to the unit. Put the alignment transforms in a text file for particle extraction.
Map particles to tomograms
There is a simple tool to map the averaged structure to the determined position and orientation of each particle in a tomogram. Available after EMAN2.3. In versions after 05/23/2019, the function is moved to the Analysis and Visualization section in the GUI.
Subtomogram Averaging -> Map particles to tomograms
Set path to be one of the spt_XX folder (not the subtlt ones).
Set iter to be the iteration you want to use from the refinement.
- Browse for one tomogram you want to map the particles to.
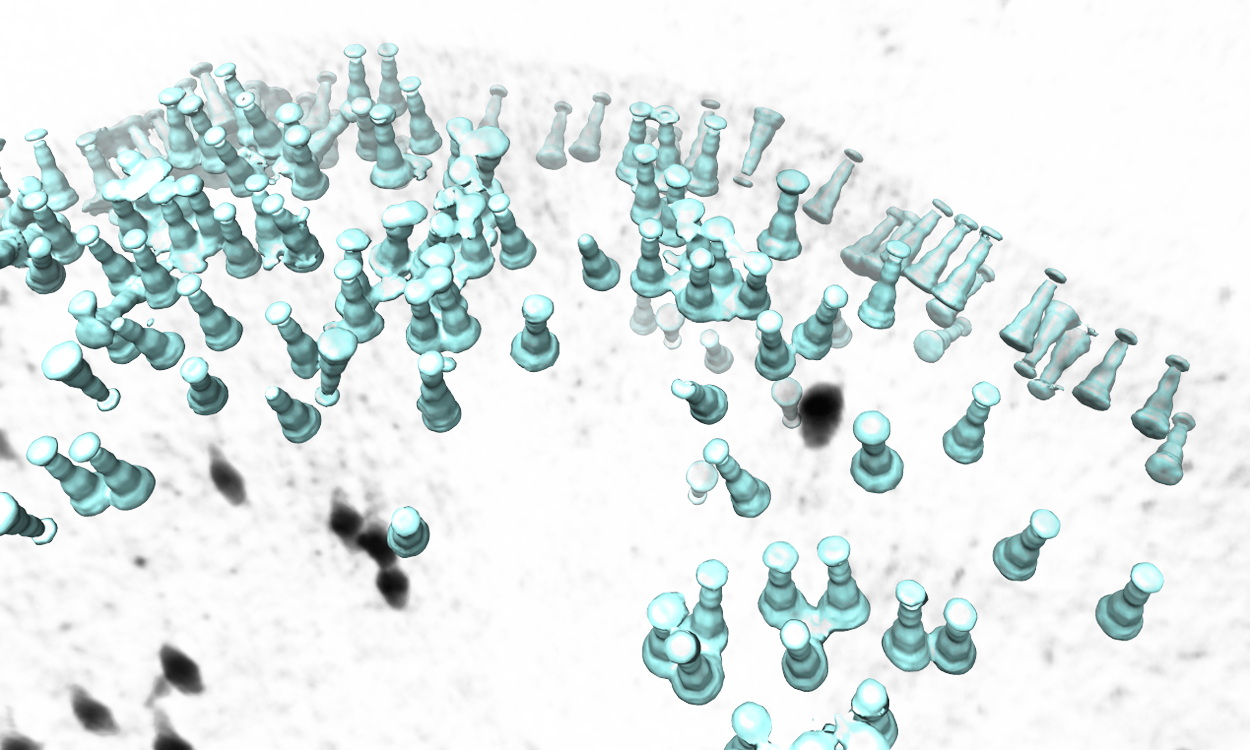
The program will then find all particles in the selected tomogram that are used in the refinement, map the averaged structure back, and produce a file called ptcls_in_tomo_xx_yy.hdf, where xx is the name of tomogram and yy is the number of iteration used. This is sometimes quite useful for objects in cellular environment (when membrane proteins are obviously upside down for example). Image rendered with Chimera.