|
Size: 53273
Comment:
|
Size: 22074
Comment:
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 1: | Line 1: |
| NOTE: This is an early version of our tomography workflow tutorial. Updates will be posted in the coming weeks describing recommended procedures for automated segmentation and subtomogram averaging. | ## page was renamed from e2tomo = EMAN2 Tomography Workflow Tutorial = |
| Line 3: | Line 4: |
| == Introduction to the tutorial == | * This tutorial is best suited for EMAN2 built after 09/27/2018. Not everything described in the tutorial was functioning yet in the 2.22 release. * This tutorial uses data from EMPIAR: [[https://www.ebi.ac.uk/pdbe/emdb/empiar/entry/10064|EMPIAR 10064]] (only the 4 mixed CTEM tilt series) * Time estimates for each step are for a well-configured tomography workstation with a high-speed disk, 64+ GB of RAM and 16+ cores. * The pixel size in the header of the files are incorrect as provided by EMPIAR. The correct Apix value (2.62) should be specified when importing the images. |
| Line 5: | Line 9: |
| EMAN2 can be used at many different levels ranging from high-level task-based workflow, to command-line utilities, to writing code in Python or C++. In this tutorial, we will be focusing primarily on the task-focused high-level Project Manager interface. This interface will help you work step-by-step through established techniques such as single particle analysis and subtomogram averaging. | == Prepare input files (~2 minutes) == * Make a new empty folder for the project and 'cd' into that folder * run '''[[http://blake.bcm.edu/emanwiki/EMAN2/Programs/e2projectmanager|e2projectmanager.py]]''' * Make sure any EMAN2 commands you run are executed from within this folder (not any subfolder) * You may use "Edit Project" from the Project menu to set default values for the project. While not required, it reduces later errors. * Make sure the workflow mode is set to "TOMO" not "SPR" |
| Line 7: | Line 16: |
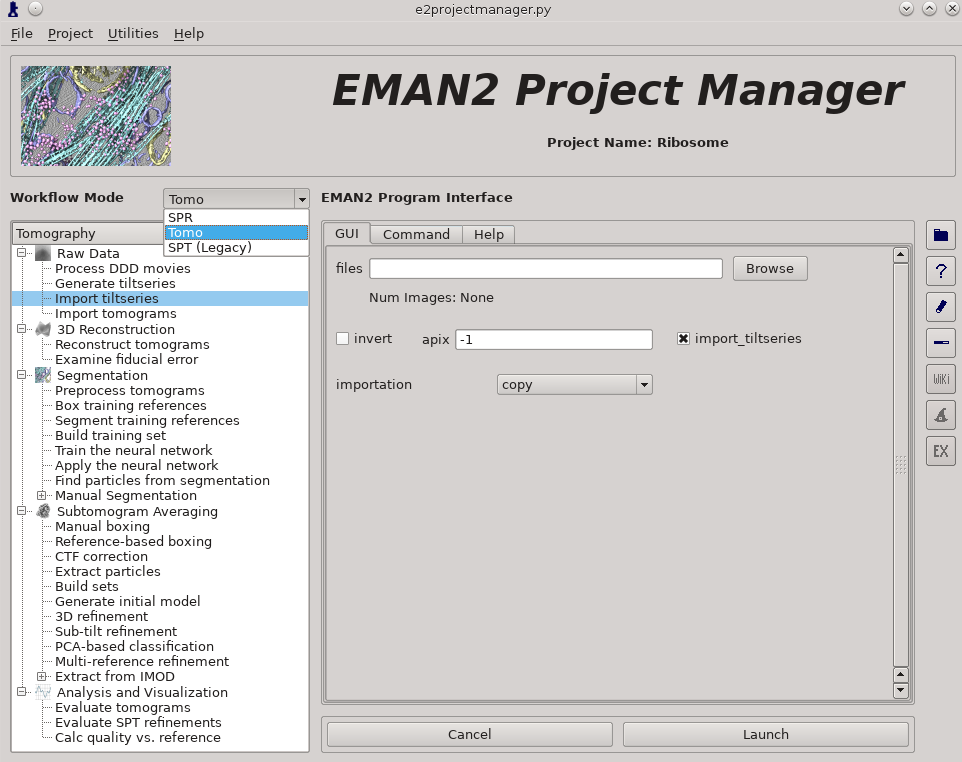
| We will be using an 80S-Ribosome data set for this tutorial obtained from EMPIAR (https://www.ebi.ac.uk/pdbe/emdb/empiar/entry/10064). The manuscript associated with this data deposition reported a final resolution of ~11Å resolution, but obtaining this result on a laptop would require significant time and it is unlikely that such hardware would possess sufficient RAM for memory-intensive processes such as 3D subtomogram refinement. Here we will operate on binned tiltseries/tomograms, reducing the attainable resolution but allowing the complete subtomogram averaging pipeline to be performed on relatively minimal hardware. | {{attachment:e2pm.png|Project Manager|width=600}} |
| Line 9: | Line 18: |
| We strongly recommend going through this tutorial using the provided data set. Once you understand how everything is supposed to work, then you can use your own data or download additional public data sets from sites like http://www.ebi.ac.uk/pdbe/emdb/empiar. | * Import tilt series using '''Raw Data -> Import tilt series''' * Select the files, and make sure '''importation''' says '''copy''' * In this step you should enter the correct A/pix for your data in the '''apix''' box. For EMPIAR10064, this is 2.62. For your own data, you need to know this number. In later steps you should be able to use -1 (default) for apix. * If your tilt series isn't a single stack file, but is many individual images instead, you will need to use '''Generate tiltseries''' to build an image stack. This is not necessary for the tutorial data. * Once the options are set, press '''Launch''' |
| Line 11: | Line 24: |
| There are several important things this tutorial does not cover: Movie mode processing: This is very data intensive, so we are skipping it for the workshop. The relevant programs are: e2ddd_external.py, e2ddd_movie.py, and e2ddd_particles.py Particle picking: There is (or will be) a separate tutorial on the website covering the new particle picking program, including the neural network-based picker. Single particle analysis: A separate tutorial covers this topic and is available on our wiki: http://blake.bcm.edu/emanwiki/EMAN2/Tutorials Detailed Command-line usage: While we will provide command line program names and a brief description of their usage at the end of this tutorial, the main focus will be our graphical interface. To obtain additional information about any of our command line programs, you can run with “--help” to see a list of available options. EMAN with Python: A fun topic, but not for the whole audience. We have some decent recordings covering this online. Please feel free to email the mailing list/google group any time for help. |
* It is critical that the filenames for your data not contain any spaces (replace with underscore) or periods (other than the final period used for the file extension). "__" (double underscore) is also reserved for describing modified versions of the same file, and should not be used in your original files. |
| Line 18: | Line 26: |
| Check the Wiki (eman2.org) for other available tutorials, some of which include videos | == Tiltseries Alignment and Tomogram Reconstruction (20 min) == Alignment of the tilt-series is performed iteratively in conjunction with tomogram reconstruction. Tomograms are not normally reconstructed at full resolution, generally limited to 1k x 1k or 2k x 2k, but the tilt-series are aligned at full resolution. For high resolution subtomogram averaging, the raw tilt-series data is used, based on coordinates from particle picking in the downsampled tomograms. On a typical workstation reconstruction takes about 4-5 minutes per tomogram. |
| Line 20: | Line 29: |
| Time estimates provided below assume you are using a single 3.1 Ghz Intel Core i7 processor. We also assume that the user has 16GB ram. EMAN2.21a Tutorial v2 July 2018 3 | For the tutorial tilt-series: * 3D Reconstruction -> Reconstruct Tomograms * check ''alltiltseries'' * alternatively you can select one or more tilt series from the ''tiltseries'' folder * check ''correctrot'' * ''tltstep'' = 2 * ''clipz'' = 64 * If you wish to look at the intermediate aligned tilt-series and other files, uncheck ''notmp'' * This is not required for the remaining steps in the tutorial, but can be used to help understand how the tomogram alignment works. This requires significant additional disk space. You may consider doing this for only one tomogram. * In each ''tomorecon_XX'' folder * ''landmark_0X.txt'' has the location of the landmarks (which may be fiducials if present) in each iteration * ''samples_0X.hdf'' shows the top and side view of those landmarks * ''ptclali_0X.hdf'' has the trace of each landmark throughout the tilt series (they should stay at the center of image all the time if the alignment is good) * ''tomo_0X.hdf'' is the reconstruction after each iteration * Launch |
| Line 22: | Line 45: |
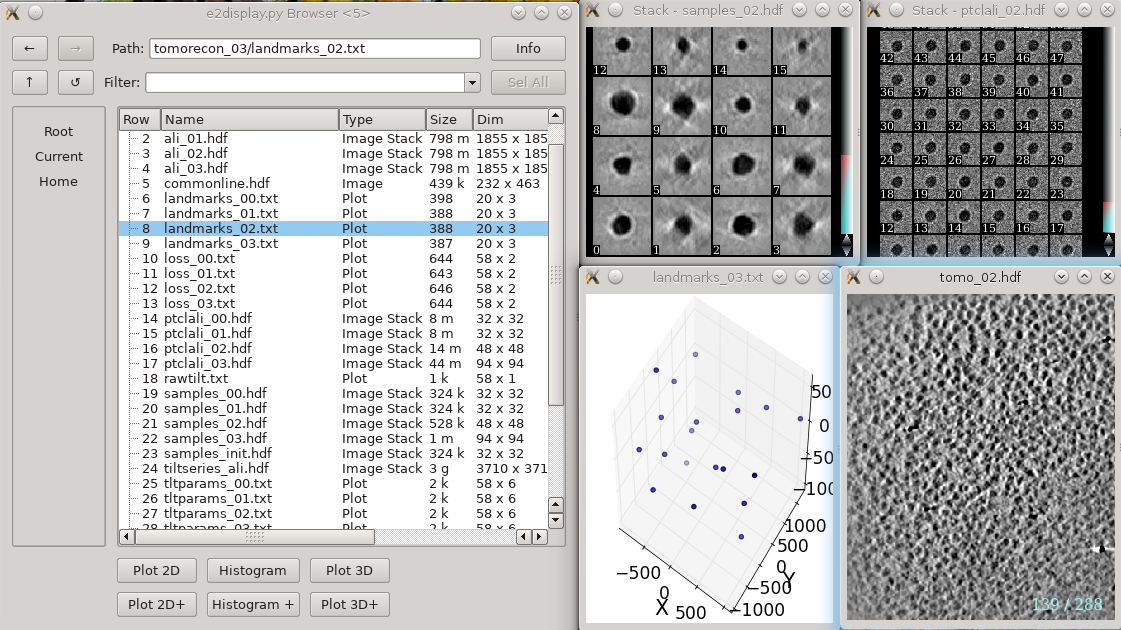
| ➡ If you are an experienced user (or an impatient one), there is a much shorter ‘quick’ version of the tutorial without all of the detailed explanations, at the end of this document. The section numbering is the same in both versions, so you can move back and forth as necessary. ie- you can go through the short version, and if you don't understand something, jump back to the detailed version for more information. The detailed section also includes many useful comments and explanations for solving problems with your own data, not just the tutorial set. | {{attachment:tomorecon.png| Tomogram reconstruction |width=600}} |
| Line 24: | Line 47: |
| Detailed tutorial | When working with your own data: * Either specify the correct ''tltstep'' if the tilt series is in order from one extreme to the other, '''or''' specify the name of a ''rawtlt'' file (as produced by serialem/IMOD). * While the program can automatically compute the orientation of the tilt axis, it is better to fill in the correct value in ''tltax'' since there is a handedness ambiguity in the tomogram if determined automatically. * In most cases, the default ''npk'' should be fine. If fiducials are present, it is not necessary to adjust this number to match the number of fiducials. The program will use any high contrast areas it finds as potential landmarks. * ''bytile'' should normally be selected, as it will normally produce better quality reconstructions at higher speed. If 2k or larger tomograms are created, memory consumption may be high, and you should check the program output for the anticipated RAM usage. * The graphical interface only permits 1k or 2k reconstruction sizes. In our experience this is normally sufficient for segmentation or particle picking. * When the sample is thin (purified protein, not cells), it is useful to check '''correctrot''' to automatically position tomograms flat in ice * It can also be helpful with thin ice to specify a '''clipz''' value to generate thinner tomograms (perhaps 64 or 96 for a 1k tomogram). |
| Line 26: | Line 56: |
| Before getting started, it's a good idea to get a feel for the relative speed of your computer (to set expectations). Run e2speedtest.py. This will give you a score telling you how fast a single processor is on your computer. If your machine has 4 cores, you multiply this number by 4 to get a relative performance value. Note, however, that some processors have a 'turbo' mode, and if you are using only 1 processor (which is what the test does), it will run faster than 1 core normally will. This can exaggerate the speedtest score by as much as 20-30%. My 2017 MacBook Pro (3.1 GHz Intel Core i7) scores ~1.3 (per core) on this test. | == CTF Estimation (10 min) == |
| Line 28: | Line 58: |
| 1. Open the EMAN2 project manager | For the tutorial tilt-series: * Subtomogram Averaging -> CTF Correction * check ''alltiltseries'' * Double check the ''voltage'' and ''cs'' * Launch |
| Line 30: | Line 64: |
| A note on project organization/management: What should you do with your raw data? | When working with your own data: * The first two options, ''dfrange'' and ''psrange'' indicate the defocus and phase shift range to search. They take the format of “start, end, step”, so “2, 5, .1” will search defocus from 2 to 5 um with a step size of 0.1. Units for phase shift is degrees. * For images taken with volta phase plate, we usually have '''dfrange''' of “0.2,2,0.1” and '''psrange''' of “60,120,2”. |
| Line 32: | Line 68: |
| We begin by unzipping the tutorial file. The compressed directory includes two directories (CTFnoise and Distortion) and a tiltseries, cryo.st that we will reconstruct using the EMAN2 tomography workflow. Open a terminal window/command prompt and move into the unzipped folder. On my computer, this looks like: | Note that this program is only estimating CTF parameters, taking tilt into account. It is not performing any phase-flipping corrections on whole tomograms. CTF correction is performed later as a per-particle process. This process requires metadata determined during tilt-series alignment, so it cannot be used with tomograms reconstructed using other software packages. |
| Line 34: | Line 70: |
| cd /home/jmbell/cryo | == Tomogram annotation (optional) == |
| Line 36: | Line 72: |
| Typically, I create a directory called “rawdata” that houses everything I want to preserve but not process. In this case, I suggest making this directory (mkdir rawdata) and moving everything in the current directory into this folder, i.e.: | {{attachment:annotation.png| 2D particle picking |width=600}} |
| Line 38: | Line 74: |
| mkdir rawdata mv ./* rawdata |
* Since the tutorial data set is purified ribosomes, this step can be skipped for the tutorial data, and you can move on to template-based particle picking. For cells or other types of complex specimens, tomogram annotation can be used to produce locations of different types of objects. |
| Line 41: | Line 76: |
| Next, from within the current folder, run “e2projectmanager.py”, which will bring up the EMAN2 project management GUI. By default, the Workflow Mode shown at the upper left of the window under the EMAN2 logo will be “SPR”. This mode provides tools for single particle analysis. We will be using the new “Tomo” mode, so click on the dropdown menu and select “Tomo”. When you do this, the workflow menu will show a new series of panels including Raw Data 3D Reconstruction Segmentation Subtomogram Averaging Analysis and Visualization |
This section is brief and is probably only useful for users already somewhat familiar with the tomogram annotation protocol. A more detailed tutorial of the subject can be found in [[http://eman2.org/Programs/tomoseg| TomoSeg]]. Note that some directory structure and user interfaces have changed in the latest version to match new tomogram workflow. * Segmentation -> Preprocess tomograms * This step is not always necessary for tomograms reconstructed in EMAN2, but may slightly improve results. * Segmentation -> Box Training References * This is a newer interface than previously used for this step. Select a few "Good" (regions containing the feature of interest) and "Bad" (regions not containing the feature of interest) boxes. * "~" and "1" on the keyboard can be used to move along the Z axis. * The new interface permits different types of features to be identified in a single session and in the same tomogram. * If the different features of interest have very different scale, it is always better to keep the box size at 64, and instead rescale the tomogram. As long as the rescaling is done using EMAN2 utilities, the program will correctly keep track of the geometry relative to the original tomogram & tilt series. * if you are doing this with the tutorial data, you would only have 2 classes of particles "ribo_good" and "ribo_bad". * When pressing ''Save'' all visible particles (box checked next to the class name) will be saved * The rest of the annotation process remain unchanged from the original tutorial, except that now, all trained neural networks and training results are saved in the ''neuralnets'' folder, and all segmented maps are in the ''segmentations'' folder. You now only specify the label of the output file instead of the full file name. * Segmentation -> Find particles from segmentation to turn segmented maps into particle coordinates. * Input both the tomogram and its corresponding segmentation, and the particles coordinates will be written into the metadata file. * Slightly tweaking the threshold parameters may yield better results. * ''featurename'' will become the label of particles generated. Those particles can be viewed in the particle picking step and processed in the following protocols. == Particle picking == {{attachment:ptclpicking.png| 3D particle picking |width=600}} Launch the boxer in '''Subtomogram averaging -> Manual boxing''' step. You can also launch it via the '''Tomogram evaluation''' step which is discussed later in this tutorial. Go through slices along z-axis using '''‘~’''' and '''‘1’''' on the keyboard and deleting boxes by holding Shift and click. The boxes are shown as circles, whose radii indicate the distance from the current slice to the center of the particles. You can rename the label of the boxes and create new types of boxes in the '''options''' window. Make sure the label is consistent for the same particle type in different tomograms, as it is used for particle extraction later. The box size can be set in the main window at the left bottom corner, in this case, 45 for the ribosomes (so the unbinned box size is 180). If you have got the particle coordinates from tomogram annotation, just take a look at the automatically generated particles here and remove some obvious bad ones. While you can save 3D particles from the GUI, there is no need to do so in this step. When you are satisfied with the result, simply close the window. You should have ~3000 particles from the 4 tomograms in the dataset. If you skipped the tomogram annotation step, we will pick a few particles here to generate an initial model first, and use the initial model as a reference for template matching. Select 30-50 particles from a tomogram, then close the boxer window. |
| Line 49: | Line 104: |
| On the right side of the project manager window, there are a series of buttons: Browser: Open the EMAN2 file browser Help: Open the e2help.py GUI that provides considerable detail about our C++ image processing utilities (aligners, reconstructors, processors, projectors, etc.). Notebook (Log): Open the EMAN2 notebook, which shows a log of processes called and the times they were launched. Task manager: Show the EMAN2 tasks/programs currently running on your computer. The last three buttons are available only for a subset of our programs. Wiki: Open additional documentation on the web Wizard: Open the project manager wizard to assist when filling in program parameters Expert mode: Show/hide additional options for a given program. |
== Particle extraction == |
| Line 59: | Line 106: |
| To keep things organized, particularly when working on multiple projects simultaneously, it is often useful to assign a unique project name to each tomography project, which will be displayed under the EMAN2 Project Manager title text. To do this, click on the project manager window and either at the top of the window or the top of your screen (depending on which operating system you’re using), click “Project”->”Edit project”. Here you can provide a project name, say “EMAN2 Tomography Tutorial”. For record keeping, we suggest also filling the particle mass, and microscope Cs, voltage, and apix (Å/pixel) values. The Å/pixel value for this tutorial dataset is 2.62. The specified parameters will be used when possible throughout the refinement process, but more importantly, collaborators will be able to access these parameters when you share your project directory, reducing the chance of processing errors due to incorrect parameter specification. | In this step, the program will extract unbinned 2D particles from tilt series, perform per particle per tilt CTF correction, then reconstruct individual 3D particles. Select '''Extract particles''' from the left panel, check '''alltomograms''', and specify the label of particle you want to extract. Make sure the label specified here corresponds to the label of particles from the particle boxer. If the box size is correct when you select particles from the GUI, you can leave '''boxsz_unbin''' as -1, so the program will keep that box size. You can adjust the value if you want to change the box size of the extracted particles. If your particles are deeply buried in other densities, using a bigger '''padtwod''' may help, but doing so may significantly increase the memory usage and slow down the process. With CTF information present, it generally does not hurt to check '''wiener''', which filters the 2D particles by SSNR before reconstructing them in 3D. If you want to generate particles without CTF correction, check '''noctf'''. By default, the generated particles will have the same label as they are named in the boxer. If you want to have multiple types of particles, for example, with and without CTF correction, you can specify a different '''newlabel''' each time you launch the program. Specify a binning factor in '''shrink''' to produce downsampled particles if your memory/storage/CPU time is limited, but it may also limit the resolution you achieve at the end. |
| Line 61: | Line 108: |
| For the EMPIAR example, check '''alltomograms''', and specify the label for the particles (either all ribosome particles or the ones for initial model); click '''Launch'''. | |
| Line 62: | Line 110: |
| Then, go to '''Build set''' in the left panel, check '''allparticles''', and click launch. This will generate particle sets, which are virtual particle stacks that consist of particles with the same label from different tomograms. | |
| Line 63: | Line 112: |
| 2. Import data | == Initial model generation == |
| Line 65: | Line 114: |
| Tomography projects have a variety of starting points. This tutorial begins from raw tiltseries; however, it’s also possible to start from raw movie frames or individual tilt images that must be pre-processed before they can be interpreted as a tiltseries. | {{attachment:initial_model.png| Initial model generation | width=600}} |
| Line 67: | Line 116: |
| While there are no specific requirements for the organization of your raw data in EMAN2, we recommend keeping a copy of your raw data on your machine and processing a separate copy. Following this recommendation, we will create a copy of the raw tiltseries from the “data” directory via the EMAN2 workflow GUI. | To build an initial model from scratch, simply go to the '''Generate initial model''' step and input the particle list. If you wish the process to be faster, set '''shrink''' to 2-4. It is not necessary to change other options. The program is parallelized, but not in a standard EMAN2 way. To use more cores, you can enter a bigger number in '''batchsize'''. This will not make the program run faster but may make it converge to the correct answer faster. Also using more particles as input won’t make it run faster or slower either, so either input the full particle set if you have them, or the 30-50 particles you pick for the initial model. If the protein is known to be symmetrical, specify the correct '''symmetry'''. The program will not actually apply the symmetry (unless you check the '''applysym''' box, which is not recommended in general), but it will align the initial model to the symmetry axis so the following steps can work. For most situations, the default number of iterations ('''niter''') of 5 is much more than needed. |
| Line 69: | Line 118: |
| Raw tiltseries | In this ribosome dataset with '''shrink''' 3, the program should converge to a good initial model before the end of the first iteration, usually within 10 minutes. Output files are written in folders called '''sptsgd_XX'''. In the output folder, the file '''output.hdf''' is the current initial model, which is updated after each batch (so 10-20 times per iteration). So it is okay to stop the program early and use the file as an initial model once it looks good enough. While it would be good to have a better early stopping criterion, given the diversity of things in the cell, we have not come up with one yet. |
| Line 71: | Line 120: |
| We define a raw tiltseries as a stack of tilt images in tilt angle order (usually negative to positive). To import raw tiltseries data, double click the “Raw Data” entry in the EMAN2 project manager workflow menu and select “Import tiltseries”. This will bring up a new display in the EMAN2 program interface where you can specify the path to the tiltseries, whether to invert contrast upon import, and whether to copy, move, or link the incoming data. | == Template matching == |
| Line 73: | Line 122: |
| Click “Browse” on the upper right of the program interface window, select “cryo.hdf”, and click “OK”. Next type 7.7 into the Apix box. Finally, make sure “copy” is selected under the importation dropdown menu and click “Launch”. | If you generated all particles with tomogram annotation already, skip this step. If not, go to '''Reference-based boxing''', click '''Browse''' for '''tomograms''' to select all tomograms, and Browse the initial model generated in the previous step as '''reference'''. Specify the label of the output particles in '''label''' and set a maximum particle number per tomogram in '''nptcl''' (in the EMPIAR example, 800 should be fine), and click '''Launch'''. |
| Line 75: | Line 124: |
| Generally we recommend that users copy their data from a separate directory (such as “rawdata”) so that we always have a backup copy of the raw data on disk in the event that files are somehow corrupted. This directory can be placed within or exist outside an EMAN2 project. However, if you prefer to move an existing copy of your data into an EMAN2 project, you can specify the “move” importation method. Alternatively, it’s also possible to manipulate the data in place using the “link” option. | After the program finishes, take a look at the particle coordinates from '''Manual boxing''' in the project manager or '''Tomogram evaluation''', and manually remove the obvious bad boxes. This may perform worse than the tomogram annotation in excluding ice contamination and fiducial, but should still be fine for purified samples in this example. Once you are satisfied with the boxes, repeat the '''Particle extraction''' step using the label for the full particle set to generate all 3D particles. |
| Line 77: | Line 126: |
| NOTE: It is essential that all imported files have the correct Å/pixel value in their header or that it is specified when running one of our import routines. If you are certain that the Å/pixel value in the header is correct, you can specify -1 in the apix box during tiltseries import. For more details on manipulating file header parameters, see the block below titled Inspecting and modifying image file header parameters. | == Subtomogram refinement == |
| Line 79: | Line 128: |
| Individual micrographs | {{attachment:refinement.png| 3D refinement | width=600}} |
| Line 81: | Line 130: |
| If starting your tomography with individual tilt images, you can import these images directly into a tiltseries by using the “e2buildstacks.py” program. This is accessible from the GUI by selecting “Generate tiltseries” under the “Raw Data” workflow menu entry. Here you will specify the images in tilt angle order (negative to positive) and type the name you wish to assign this tiltseries. Before clicking “Launch”, be sure the “tilts” box is checked. The resulting tiltseries will be stored in the project “tiltseries” directory. If this directory does not already exist, the program will create it automatically. | Click '''3D refinement''' from the left panel, and input both the particle set and the initial model generated from the last step as a reference. If there is a symmetry of the protein, make sure it is aligned to the symmetry axis before specifying the correct symmetry. If you are willing to split the even/odd set of particles and do a “gold-standard” refinement, specify a resolution number (usually 30-50) in '''goldstandard''', so information beyond that resolution will be randomized independently in the reference for even and odd set. While it is good to have a reasonable '''mass''' for the molecular weight of protein (in kDa) and '''tarres''' for the target resolution, leaving them as default usually does not hurt. If you have a known structure factor in a .txt file, (you can compute it from a known structure via [[http://blake.bcm.edu/emanwiki/EMAN2/Programs/e2proc3d|e2proc3d.py]]), specify it in '''setsf'''. '''localfilter''' will filter the averaged map by local resolution, which is especially useful when looking at things in cells where parts of proteins can be very flexible. This is almost always good to check when you want to push toward high resoluion. '''pkeep''' controls the fraction of particles that go into the final average. If you know there are many bad particles in the dataset, setting it to be a smaller number may help. Enter the number of threads you want to use in the '''thread''' option. Finally, click '''Launch''' and wait. For this dataset, it can take a few hours on a decent workstation. The results can be seen in the '''spt_XX''' folder. In the folder, '''threed_XX.hdf''' files are the main output map after each iteration, and '''fsc_masked/unmasked/masktight_XX.txt''' files are the FSC curves between even/odd half set under different masking. You should be able to get to 12-15Å resolution (cutoff 0.143) at this step using this dataset. |
| Line 83: | Line 132: |
| DDD frames | == Subtilt refinement == |
| Line 85: | Line 134: |
| If starting from raw DDD frames, you may or may not have an mdoc file containing relevant tilt angle and file name information used to combine aligned frames into a tiltseries. In cases where such a document is unavailable, one can align the frames individually using the e2ddd_external.py program, available through the GUI under “Raw Data” in the workflow menu. To use this program, simply select “Process DDD movies” and provide a list of the movies you want to align in tilt angle order. | {{attachment:subtlt_dir.png| Subtilt refinement directory |width=600}} |
| Line 87: | Line 136: |
| If you have an mdoc file, the process is significantly easier. In the “Process DDD movies” program interface window, begin by clicking “Browse” next to “mdoc” file box and select the relevant mdoc file for the movie data you wish to align and convert into a tiltserles. Next click “Browse” in the “input” file input box and select either a list of movies in arbitrary order or a directory containing the movies to be aligned. When launched, the program “e2ddd_external.py” will organize, align, and save the tilt images in tilt angle order according to the contents of the specified mdoc file. | Once the subtomogram refinement finishes, check the final map and FSC curves. In this dataset, you should be able to achieve a resolution of 13-15Å. Now we can refine the orientation of each individual subtilt, i.e. 2D particles from raw tilt series that are reconstructed into to the 3D particles, and push the resolution of the averaged map. |
| Line 89: | Line 138: |
| During alignment, either IMOD’s alignframes routine or MotionCor2 will be used. Note that for these programs to work, they will need to be installed and available in your PATH environment variable. Correct installation instructions for these programs/packages are available from their respective developers/distribution platforms. | Click '''Sub-tilt refinement''', choose the folder of the last subtomogram refinement and launch the program. You will need to specify the '''path''' to the spt_XX directory containing the last completed subtomogram refinement (typically just “spt_00” for example). Additionally, specify the '''iter''' you want to use as a starting point for sub-tilt refinement. If “-1” is specified, the program will attempt to locate the last complete iteration. |
| Line 91: | Line 140: |
| Optionally, you may specify dark and gain reference movies in the corresponding file boxes, and we offer some basic options for the two alignment routines. For more advanced usage, we suggest that users runs these programs from the command line. Including these in our GUI is solely for the sake of convenience. | The default parameters should be generally fine for this dataset, though you may need to alter the '''parallel''' and '''threads''' options to use the number of CPU threads available on your computer. The niters value corresponds to the number of iterations of sub-tilt refinement you wish to perform. '''keep''' controls the fraction of particles that goes into the final map. If you are certain that tilt images beyond a certain angle (for example, 45 degrees) are radiation damaged, you can put 45 in '''maxalt''', and specify a larger keep number. Otherwise, just use '''keep''' 0.5, so the program will judge the quality of subtilt images by their correlation to the averaged map and exclude worst 50% 2D particles. |
| Line 93: | Line 142: |
| The final output of e2ddd_external.py when run within the “Tomo” workflow is an unaligned tiltseries, which will be stored in the “tiltseries” project folder. | == Tomogram evaluation == |
| Line 95: | Line 144: |
| NOTE: When aligning DDD movie data using e2ddd_external.py via the command line or project manager interface, it is essential to verify that all imported files have the correct Å/pixel value in their header. For instructions on how to inspect/modify image file header parameters such as Å/pixel, see the block below. | {{attachment:tomo_evaluation.png| Tomogram evaluation |width=600}} |
| Line 97: | Line 146: |
| Inspecting and modifying image file header parameters: | This is a tool that helps you visualize your tomograms with their corresponding metadata, and launch other programs from it. It can be found via '''Analysis and visualization -> Evaluate tomograms'''. This can be used at any point of the workflow after tomogram reconstruction. |
| Line 99: | Line 148: |
| To inspect a file’s header parameters (e.g. apix_x, apix_y, etc.), you can either use the EMAN2 file browser or command line. | On the left is a list of tomograms in the project. Clicking the header of each column will sort the table by that attribute. '''#box''' is the number of boxes in the tomogram, '''loss''' is the average fiducial error in nm, and '''defocus''' is the average defocus of the tilt series. Do not be scared by large '''loss''' values here. Although the relative value of different tomograms (aligned with the same parameters) in the same project are correlated with tiltseries quality, the exact value here is not as meaningful. You can still get a subnanometer resolution subtomogram average from tilt series with a loss larger than 5 nm. |
| Line 101: | Line 150: |
| If you prefer a graphical interface, click on the folder icon from the project manager or run e2display.py via the command line. Next, navigate to the tiltseries directory and single click on an imported tiltseries. Next click the “info” button at the upper right to inspect the header parameters. | On the right, the image on the top shows the center slice of the tomogram. The '''Show2D''' button shows the selected tomogram in slices, '''!ShowTilts''' shows the corresponding raw tilt series, and '''Boxer''' calls the 3D boxer. '''!PlotLoss''' will plot the fiducial error per each tilt, and '''!PlotCtf''' plot the defocus and phase shift at the center of each tilt image. '''Tiltparams''' is a bit more complicated. It plots a point list with 6 columns and a number of rows corresponding to the images in the selected tilt series. These are the alignment parameters for the tilt series. The columns represent tilt ID, translation along x and y-axis, tilt angle around y, x and z-axis correspondingly. You can adjust '''X Col''' and '''Y Col''' in the plot control panel (middle click the plot) to change the display. The first panel below the buttons are the types of particle and their numbers in the dataset. Check and uncheck the boxes will affect the number displayed in '''#box''' column on the left. The last box is reserved for comments for each tomogram. You can fill in any comments you have for the selected tomogram and it will be saved with other metadata of the tomogram for future references. |
| Line 103: | Line 152: |
| Alternatively, from the command line, it is possible to obtain header parameters by running the command “e2iminfo.py filename.ext -H”, which will print the contents of the header to the terminal window. In either case, you should examine the “apix_x”, “apix_y” header parameters and ensure that these values are consistent with the magnification used during data collection and binning applied before/during tiltseries importation. | == Refinement evaluation == |
| Line 105: | Line 154: |
| If these values are incorrect and need to be modified, this task can be accomplished using the command line program, “e2procheader.py”. Specifically, to change the apix value to a preferred value, say 7.699, you would run the following command from within the tiltseries directory (on an already imported tiltseries): e2procheader.py --input tiltseries.hdf --output tiltseries.hdf --stem apix --stemval 7.699. | {{attachment:refinement_evaluation.png| Refinement evaluation |width=600}} |
| Line 107: | Line 156: |
| 3. Reconstruct tiltseries Once tiltseries data is imported into the EMAN2 project directory, we can proceed with tiltseries alignment and 3D reconstruction. This is a fully automated procedure in EMAN2 that begins with a coarse, cross-correlation alignment of tiltseries followed by rounds of iterative refinement. Each iteration consists of generating a tomogram, picking high-contrast landmarks in 3D (rather than relying solely on gold fiducials), mapping landmark coordinates to 2D tilt images, refining the coordinates of those landmarks, and then refining the alignment parameters of each image in the tiltseries. We repeat this process with different levels of binning to focus alignment on low resolution features first-and-foremost. The final result is a reconstructed tomogram that is either 1024x1024x256 (default) or 2048x2048x512 depending on which options are specified. It’s worth noting that unlike most tomogram reconstruction software that uses SIRT or back-projection algorithms, EMAN2 performs reconstructions using direct Fourier methods, similar to how we perform reconstructions in single particle analysis. To perform a tiltseries alignment and tomographic reconstruction using the project manager interface, begin by double clicking on the “3D Reconstruction” workflow menu item and select “Reconstruct tomograms.” Next, click “Browse” at the upper right of the program interface window and select all the tiltseries you wish to reconstruct. In this case, you should see the “cryo.hdf” tiltseries you imported in the last step. Single click on this file (“cryo.hdf”) and click “OK”, which will close the current window. In EMAN2, it is not necessary to specify a rawtlt file. Instead, we assume that images in a specified tiltseries are in the correct (tilt angle) order with no missing images. If this is the case, users need only to specify a tilt step (tltstep) and the index of the 0° tilt image (zeroid). For this dataset, the middle image is the 0° image (zeroid=-1), and the angle increment between tilts is 2° (tltstep=2), so the default parameters will work. However, in cases when your tilt step is different, is essential that you specify an accurate value in the tltstep parameter box. While the default parameters should work well for this tutorial dataset, we recommend that you modify the number of threads at the bottom of the program interface window to correspond to the number of cores on your computer. Note that on a core i7 processor, I can run 6-8 threads without problems. On a core i3 or i5, I would not run more than 4. For more details about each of the options available for this program, you can hover over the parameter in the program interface window or run “e2tomogram.py --help” from the command line. Once all parameters are set, click “Launch” to begin alignment and reconstruction. A complete reconstruction on a full 4k x 4k dataset usually takes ~8-12 minutes on 12 threads (depending on your hardware). In comparison, the “cryo.hdf” tiltseries is only 2k x 2k and should require only about ~3.6 minutes to reconstruct on a high-end laptop. While running, the program writes alignment information to ‘info/xx_info.json’, including ali_loss : average residue error for each tilt, in nm tlt_file : tilt series file input for the reconstruction tlt_params : transform parameters for each tilt. 5 columns represent translation x, translation y, tilt axis rotation, tilt angle, off axis tilt angle. If run with the “notmp” box left empty, a new folder called ‘tomorecon_xx’ will be created, containing the following intermediate files: ali_xx.hdf : aligned tilt series with 3D transform in ‘xform.projection’ in their header landmark_xx.txt : 3D location of the landmark used. loss_xx.txt : average residue error for each tilt, in nm ptclali_xx.hdf : per particle landmark tracking results in 2D. In the header of images, ‘nid’ is the index of tilt series, ‘pid’ is the index of the landmark, and ‘score’ is the (x,y) translation alignment of the particle. samples_xx.hdf : top and side (x-z plane) view of each landmark in 3D. A good way to evaluate the refinement is to see how round are the side views.. tltparams_xx.txt : transform parameters for each tilt after each iteration tomo_xx.hdf : bin8 tomogram reconstruction after each iteration. Note that we shrink by minimum instead of mean value when we make bin8 tilt series for reconstruction and landmark search, so small high contrast landmarks are not averaged out. So these tomogram will look a bit strange as dark things are often larger than expected (the actual final tomogram output still uses mean shrinked tilt series).. Once finished, bin x4 versions tomograms are written to ‘tomograms/xx__bin4.hdf’. 4. Evaluate reconstructions Now that your tomogram(s) have been reconstructed, it’s a good idea to take a look at the results before performing subsequent processing and analysis. While the default reconstruction parameters have successfully reconstructed a wide variety of tomograms, it may be necessary to perform additional rounds of reconstruction to enhance contrast or reduce artifacts through improved tilt image alignment. To investigate the quality of your reconstructions, we recommend using the e2tomo_eval.py program, which is available from the GUI by first double clicking “Analysis and Visualization”, selecting “Evaluate tomograms”, and clicking “Launch.” Once launched, the main window will appear. In the table on the left, you’ll find a list of each tomogram reconstructed in this project, the number of subtomogram boxes stored for each tomogram, and a “loss” value, which corresponds approximately to the alignment error in nanometers. On the right is a blank image display that will show a central x-y slice of a reconstruction when one is selected by clicking a row in the table on the left. Below this image display are a series of buttons. The “Show2D” button will open a slice-wise display of the selected tomogram. The “Boxer” button will open the e2spt_boxer22.py program used to box particles in 3D for later extraction. This is a cleaned up version of previous ‘e2spt_boxer’ with the support of the current metadata functionality of boxing multiple types of particles. More details about this process will be provided in a later step focusing on subtomogram boxing in EMAN2. The “Refresh” button should be pressed anytime project parameters change while this program is open. For example, if new particles are boxed, pressing “Refresh” will update the #box column values to include your selected/removed particles. The “TiltParams” button will bring up a plot window to display alignment data for each tilt image. The display will show columns 0 and 1 of the tilt parameters matrix corresponding to the x and y-translation of each image in the tiltseries; however, there are a total of 5 columns. In order, these correspond to x-translation (tx), y-translation (ty), in-plane rotation (alpha), tilt about the y-axis (ytilt), and tilt about the x-axis (xtilt). To switch which columns are plotted along the X and Y axes, simply middle-click the plot and scroll through the “X Col” and “Y Col” boxes in the inspector window that appears. The “PlotLoss” button will bring up a 2D plot window showing the values of the “loss” function during tiltseries alignment. Typically, the deeper the trough you observe, the better the tomogram reconstruction will be. While this plot is useful for debugging certain alignment problems, we do not recommend attempting to interpret this beyond choosing whether to repeat the reconstruction process using different parameters. Finally, the “PlotCtf” button will bring up a 2D plot window showing the defocus for each tilt image by default. When the plot is middle-clicked, an inspector window will appear in which different columns can be selected. 5. Tomogram annotation/segmentation If you wish to annotate the reconstructed tomograms, EMAN2 offers automated procedures to accomplish this. For details, see the tutorial at the following link: http://blake.bcm.edu/emanwiki/EMAN2/Programs/tomoseg. Note that recent changes to the EMAN2 tomography workflow have replaced the “TomoSeg” dropdown menu item with “Tomo”. However, you can find the same segmentation tools desribed in the tutorial under the “Segmentation” Workflow Menu item after selecting the “Tomo” workflow in the EMAN2 project manager. For more details about the automated EMAN2 segmentation protocol, see: Chen, M., Dai, W., Sun, S. Y., Jonasch, D., He, C. Y., Schmid, M. F., Chiu, W., and Ludtke, S. J. (2017), Convolutional Neural Networks for Automated Annotation of Cellular Cryo-electron Tomograms. Nature Methods. 14: 983-98 https://www.nature.com/protocolexchange/protocols/6171 While EMAN2 does not offer solely manual segmentation utilities, we do offer some semi-automated routines for drawing curves and contours. Currently, these are made available through two programs, namely e2tomo_drawcurve.py and e2tomo_drawcontour.py; however, we anticipate incorporating them into a single program in the future alongside other semi-automated annotation tools. e2tomo_drawcurve.py is a simple GUI tool for manually tracing curves in a reconstructed tomogram. In cases when you only have a few fibers to segment in a tomogram, this sort of semi-automated segmentation can actually be easier than fully automated methods! This approach actually has a built-in simple travel salesman problem (TSP) solver, so the user does not have to add points sequentially. Instead, one can anchor two ends of a fiber and add points between them to improve the overlap of the curve and the feature(s) of interest by building the minimal path that visits all selected points. To use this program, Mouse click on the terminus of one feature to add the first point. Next, hover above opposite terminus and and click again. To add a new contour hold “ctrl” and click. To remove a point, hold “shift” and click. The program will save points as a text file or pdb file depending on the user’s preference. EMAN2 also offers separate scripts to interpolate the points, extract particles along the curve as subvolumes and refine the position of points by alignment. Similar to ‘e2tomo_drawcurve.py’, e2tomo_drawcontour.py allows users to annotate closed contours in a semi-automated manner. It also has a built-in TSP solver for building the minimum loop and uses a simple SNAKE algorithm for fitting the contour from the previous slice to the next slice by simply pressing the “shift” key on an adjacent slice. Currently, annotation output is generated as a point cloud text file, which can be converted into a density map and displayed in rendering programs such as Chimera. 7. Subtomogram boxing EMAN2 provides users with a GUI for manually boxing subtomogram volumes from 3D tomographic reconstructions; however, we also provide tools for automated picking via reference and clipping boxes from annotations obtained using our automated segmentation workflow. Note that regardless of how boxes are picked, their coordinates are stored in json files corresponding to each tomogram in a project, which are kept in the “info” directory that houses all project metadata. To view box parameters, we recommend opening a particular tomogram using e2spt_boxer22.py, which is accessible via the e2tomo_eval.py GUI or via the EMAN2 project manager GUI under “Manual boxing” within the “Subtomogram averaging” workflow menu item. Manual Boxing Particularly when analyzing in situ datasets, there are often more than one type of particle, and the same dataset can be used to study many things. Our latest boxer GUI is designed to handle multiple particle labels when boxing manually or with a reference or prior segmentation. This allows users to explore multiple protein targets within the same EMAN2 tomography project. To manually box particles using the latest boxer program, double click on “Subtomogram averaging” in the workflow menu and select manual boxing. In the program interface window, click “Browse”, select “cryo.hdf”, and click “OK.” Next, click “Launch.” This will open the e2spt_boxer22.py widget. In cases where you have multiple tomograms to box manually, we recommend accessing the boxer from the e2tomo_eval.py program instead, as it is more convenient. Three windows will appear when the boxer GUI is launched. The main window shows a large XY view of the specified tomogram. The left column shows the current YZ-slice and the lower image displays the current XZ-slice, which can be manipulated by dragging the slider on the far right. The current box size can be changed in the Box Size input box in the lower left corner. Additionally, multiple slices can be averaged using the integer scroll box. To average all slices, click the “MaxProj” button. Occasionally, it is helpful to filter the slices to exaggerate particle features. This can be done by dragging the Filt slider bar. Magnification is controlled using the Sca slider bar. When you click on a particle in the tomogram, a box will appear. To move a box, click and hold, then drag it to a new location. To erase a box, hold shift and click on the box you wish to remove. Boxed particles will appear in the “Particles List” window. They can be easily removed by holding “shift” and clicking particles in this window. This is particularly helpful when trying to remove contaminants after performing automated boxing. The “Options” window has two main sections. The top bar turns on and off a particle box eraser. When this box is checked, your mouse clicks and drags will erase particles falling within the “Radius” in the Options window. Below this is a “Sets” tab. Here users can assign names to sets, create new sets, delete sets, and save sets. Once you have boxed various features of interest. Simply close the e2spt_boxer22.py program and all particles will remain in the project metadata for subsequent extraction. Note: If you box particles in a reconstruction and perform a second reconstruction that alters the alignment parameters, particle boxes may no longer correspond to the tomogram. In such a case it is necessary to re-box all particles (recommended) or manually manipulate the box coordinates in the metadata (NOT recommended). Reference-based Boxing Often, rather than manually selecting particles by hand, it is more efficient to detect features of interest by cross-correlating a reference map with the 3D tomogram reconstruction to identify candidate particle coordinates. When dealing with purified samples, this approach is often faster than CNN-based boxing but can produce more false-positives. To perform reference-based boxing from the EMAN2 project manager, click “Reference-based boxing” under the “Subtomogram averaging” workflow menu item. Next, click “Browse” next to the tomograms file box, select “cryo.hdf”, and click “ok.” Next click “Browse” next to the reference file box, select the 3D reference/template map you wish to use for boxing, and click “OK.” If you wish to uniquely identify particles boxed with this reference and this parameter set, type a simple, unique identifier into the “label” box (such as “ribo1”) and click “Launch.” Note that the name of boxed particles can be easily changed later via the e2spt_boxer22.py program. However, if particles are automatically boxed with non-optimal parameters and the automated procedure must be repeated, it is sometimes helpful to have the original boxing results to compare with can be helpful to hold onto the original. By labeling each instance of the reference based picking uniquely, we maintain a record of how the boxer performed given different parameters. Once we’re happy with the reference-based picking results, we can rename the particle set accordingly (i.e. “ribo” instead of “riboN”). If the default parameters do not provide satisfactory results, the following parameters can be manipulated: delta: delta angle for generating rotated references. dthr: minimum distance between particles vthr: n-sigma value threshold for particles from the output correlation. Default is 2. nptcl: maximum number of particles. Segmentation-based Boxing If you have segmented a tomogram in EMAN2, the segmentation can be used directly to produce boxes for subtomogram averaging. However, currently this operation is only supported from the command line. First, to generate particle coordinates from the segmentation output, run: extractptclfromseg.py <segmentation output> <input tomogram for segmentation> --thresh <intensity threshold in the segmentation output>. Note that the second argument has to be the tomogram you provided for the 'apply to tomogram' step in the Tomoseg workflow. If you are segmenting continuous features (e.g. microtubules) and there are no individual particles, run: extractptclfromseg.py <segmentation output> <input tomogram for segmentation> --thresh <intensity threshold in the segmentation output> --random <number of particles>. This will seed particle coordinates at random points where the intensity segmentation output is above the threshold value. The program will write particle coordinates to standard EMAN2 particle metadata corresponding to the input tomogram, same as manual particle boxing. So the extracted particles can be viewed in the tomogram using: e2spt_boxer22.py <input tomogram for segmentation> You can manually add or remove particles in the GUI. Once you are satisfied, you can generate particles from the e2spt_boxer GUI. If you are confident in the automated segmentation and do not want to go through the spt_boxer step, or you want to extract particles from the raw unbinned tomogram, run: extractptclfromseg.py <raw tomogram> <input tomogram for segmentation> --genptcls <output particle stack name> --boxsz <box size>. The first argument can be any binned or filtered version of the tomogram and the second argument has to be the same as the argument in the previous extractptclfromseg command. If you have a binned particle stack from somewhere else (like e2spt_boxer), this program also allows you to extract the same particles from the unbinned raw tomogram using extractptclfromseg.py <raw tomogram> <input particle stack> --genptcls <output particle stack name> --boxsz <box size> 8. Measure CTF (determine per-particle defoci) EMAN2 can perform CTF correction on an individual particle level using the program e2spt_tomoctf.py, which is vital to obtaining high resolution beyond the spatial frequency of the first CTF zero. The important thing to know is that we use low-tilt, high-signal information to constrain the defocus range searched when measuring the defocus of each particle locally within each tilt in a tiltseries. EMAN2 can also handle phase plate data, but that is beyond the scope of this tutorial. To perform per-particle, per-tilt CTF correction, navigate to the “Subtomogram averaging” workflow menu item and select “CTF correction.” Click “Browse” and select 1 or more tiltseries. In this case, select “cryo.hdf”. Next, select the defocus range to search by inputting values in the “dfrange” box (minimum defocus, maximum defocus, defocus step). Specify the voltage of the microscope used as well as its Cs value and click “Launch.” CTF parameters will be stored in json metadata files contained within the project “info” directory. 9. Extract subtomograms Once particles have been boxed and (optionally) CTF corrected, it is time to extract them using the e2spt_extract.py program. To perform particle extraction via the project manager interface, click “Extract particles” in the workflow window, click “Browse” next to the tomograms file box, and select “cryo.hdf”. Specify the box size you wish to use via the “boxsz” parameter. Here I am using 32. If label is not specified, all labeled particle sets will be extracted. If you did not perform CTF correction, check the “noctf” box. Otherwise, we recommend performing Wiener filtering, so check the “wiener” box. Finally specify the number of threads you wish to use for this process. On my core i7 system, I am choosing 8. Click “Launch.” While running, this program will generate bin4 particles from tomograms via e2spt_boxer.py. Additionally, unbinned 3D particles will be generated using e2spt_subtilt.py. This will take coordinates from bin4 particles and map them back to raw tilt series and generate 2D sub-tilts for each particle, and reconstruct 3D subtomograms. If CTF information exists in a tomogram’s metadata, the program will use that information to calculate the defocus of each particle, and flip the phase of 2D sub-tilt images before making 3D volumes. Additional options for more advanced usage include: padby : padding factor when extracting sub-tilt images. Default is 2. The program will also pad by an extra 1.5x when doing 3D reconstruction. It seems that padding is never enough.. maxtilt : maximum tilt angle to include in the reconstruction. This is slightly different from the same parameter in other programs since it affect the raw 3D particle that goes into alignment. But it still seems to be useless. Output sub-tilt images (particles extracted from 2D tilt images) are written to ‘particles/’ and 3D subtomograms are placed in ‘particles3d/’, under the same name as the input, but without the ‘binX’ tag. 10. Build sets When using particles from multiple tomograms, it is convenient to reference them as a single particle stack. To accomplish this, we create list files using e2spt_buildsets.py. From the GUI, we perform this process by clicking on the “Build sets” workflow menu item. Next click “Browse” and select the particle stacks from each reconstructed tomogram. Note, however, that you should not include particle set generated during segmentation. Once all files are selected, click “OK”, check “allparticles” in the program interface window, and click “Launch”. Almost instantaneously, this program will create a “sets” directory” and generate a single list file for each particle type assigned a label during subtomogram boxing. 11. Generate initial model(s) Reference-free initial modeling is critical for discovering unknown proteins in cell. EMAN2 utilizes a stochastic gradient descent (SGD) approach to perform reference-based and reference-free initial model generation for subtomogram averaging. The process starts by averaging particles at random orientation and gradually converges upon an initial model. Convergence of PSII arrays on thylakoid membranes. To perform initial modeling via the EMAN2 project manager GUI, click on “Generate initial model” in the workflow menu and click “Browse.” Next, select the set containing the relevant particles you wish to use to create an initial model (i.e. sets/particles_00.lst). If you wish to perform reference-based initial modeling, simply specify a reference using “ref”. Also, if you suspect some symmetry, it can be specified using the “sym” and “applysym” options shown in the program interface. Otherwise, we recommend using the default values initially, so click “Launch.” Additional options are available for special cases: filterto : filter maps to a certain resolution learnrate : increment of map per iteration. We have noticed that increasing this value for higher symmetry objects helps improve convergence. batchsize : number of particles in each batch. Since the multithreading in the program is based on the batches, it will go through all particles faster if batchsize is larger. However, changing this may also impact convergence. Initial models are saved in folders named ‘sptsgd_XX’ which contain 3 files. ‘Ref.hdf’ or ‘input_model.hdf’ is the initial model from random averaging or user input. ‘Output.hdf’ is the current output, which is updated after each batch (so it is always the latest model if one terminate the program before it finishes). ‘Tmpout.hdf’ is a stack of output per batch. 12. “Gold standard” 3D subtomogram refinement Once an initial model is obtained, our “gold standard” subtomogram alignment and averaging routine can be used to produce an initial reconstruction. Specifically, after dividing our data into even and odd halves as has become standard practice, we perform missing-wedge aware subtomogram alignment and averaging. Results are post-processed and filtered by the local or global even/odd-FSC, and a final map is generated after each iteration. The final result is a FSC-filtered map. To perform this series of tasks, you can either run e2spt_refine.py from the command line or use the following steps to access this program from the EMAN2 project manager. Begin by navigating to “Subtomogram Averaging” in the EMAN2 workflow menu and click “3D refinement”. Next to the particles file box, click “Browse”, select the “Ribosome.lst” particle set, and click “OK”. Beside to the reference file box, click “Browse”, select the initial model you generated previously, and click “OK”. In the “niter” box, type “4”, in the “sym” box, type “c1”, in the “mass” box, type “3200”, and in the “tarres” box, type “10”. In the “threads” box, specify the number of cores to use when running this process. Once you are done, click “Launch” to begin iterative 3D subtomogram alignment. Internally, e2spt_refine.py will scale and clip the reference to the size of particles and run a specified number of rounds of ‘e2spt_align.py’,‘e2spt_average.py’, and ‘e2refine_postpocess.py’. Required options (entered as per the instructions above) include: niter : number of iterations. Default is 5 threads : only threading. mass : mass of particle for normalization in ‘e2refine_postprocess’. tarres : target resolution used in ‘e2refine_postprocess’. Additional options for more advanced usage include: goldstandard : followed by a resolution number for phase randomization. setsf : in case there is a structure factor text file. Otherwise, no structure factor will be applied. pkeep : fraction of particles to keep. It will compute a ‘--simthr’ for ‘e2spt_average’ to keep the fraction in each iteration. mask : how to mask after each iteration. It accept mask processor like ‘mask.soft:outer_radius=-1’ or a file name of the mask. maxtilt : max tilt angle for ‘e2spt_average’. 13. Sub-tilt refinement One of the trends leading the field of single particle tomography toward and even beyond subnanometer resolution is the use of per-particle, per-tilt methods. In EMAN2, we facilitate per-particle per-tilt CTF correction, per-particle per-tilt alignment, and bad-tilt exclusion within particles. By correcting for these distortions via per-particle per-tilt alignment methods, we obtain higher fidelity subtomograms that yield improved resolution when averaged with other refined subvolumes. In the workflow menu under “Analysis and Visualization”, click “Sub-tilt refinement” Specify the path to the “spt_XX” directory, corresponding to the final 3D refinement In the “iter” box, type 3 to run for 3 iterations. In the “threads” box, specify the number of cores to use when running this process Check the “dopostp” box Click “Launch” e2spt_tiltrefine.py takes the results from a spt alignment and use it to refine the alignment of 2D particles in the sub-tilt images. The alignment is done in the gold-standard way, using ‘threed_xx_even.hdf’ and ‘threed_xx_odd.hdf’ as reference. It takes the transform from tilt series alignment and subtomogram alignment to compute the initial alignment of the sub-tilt, and only does a ‘refine’ alignment from that so it should not be far off. Since we have the correlation from the per sub-tilt alignment, we weight the sub-tilt base on the correlation and exclude the worst (now 50% sub-tilt) instead of simply excluding high angle tilts. In experiments the correlation score and tilt angle seems to be highly correlated but excluding images based on correlation gives better results than that based on tilt angle. spt_tiltrefine.py --path <existing spt_xx path> --iter <current iteration in spt_xx> --path : a spt_xx path that has the output from ‘e2spt_align.py’, ‘e2spt_average.py’ and optionally ‘e2refine_postprocess.py’. --padby : padding factor for reconstruction of subtomograms. Default is 2. --keep : fraction of sub-tilts to keep. Default is 0.5 --maxres : maximum resolution for comparison in the alignment. When running from ‘spt_refine.py’, it will use the 0.3 cutoff of FSC --unmask : use the unmasked maps as reference. --maxalt : exclude high tilt images Output files includes: threed_xx_ali.hdf : 3D map output . threed_xx_ali_even/odd.hdf : even/odd sub-maps fsc_xx_ali.txt : FSC curve after refinement. It will also rename the existing FSC of this iteration to fsc_xx_raw.txt since e2refine_postprocess overwrites.. Ribosome (EMPIAR-10064) maps and FSCs before and after 1 round of sub-tilt refinement. 14. Evaluare SPT refinements We have implemented a program called e2spt_eval.py that allows users to assess all SPT refinements performed within a given project. To run this program from the project manager, double click “Analysis and visualization” in the Workflow menu and select “Evaluate SPT refinements.” Then click “Launch.” The window that appears will show a large table on the left with rows corresponding to each SPT refinement performed. Clicking a row will show a 3D view of the map produced during the final iteration of the selected refinement. If you click “ShowBrowser,” a browser window will appear that changes to the currently selected refinement directory. The “PlotParams” button will bring up a 2D plot where you can explore the per-particle alignment parameters for each particle used in a given refinement. Here you can also examine iteration to iteration values to explore properties such as convergence or parameter-dependent clustering of particle data (as would be seen in cases with strong preferred orientations, or possibly in the presence of significant ice contamination). Finally, the “PlotFSCs” button will bring up a window showing the FSC calculated for each iteration during which post processing was performed. This is helpful when examining convergence asa well as determining the final gold standard resolution of a given SPT refinement. 15. Addressing heterogeneity 15a. Multi reference refinement In the workflow menu under “Analysis and Visualization”, click “Multi-reference refinement”. Next to the “particles” box, click “Browse”. Select the set containing your ribosome particles (“Ribosome.lst”) and click “OK”. Specify reference maps corresponding to the various states you hope to draw out of the data. If performing a focused classification of particles, specify a mask in the “mask” file box. Use “Browse” to search for this file. In the “threads” box, specify the number of cores to use when running this process. In the “tarres” box, specify the resolution target for multi-model refinement. In the “mass” box, type “3200”. Click “Launch”. 15b. Focused classification 15c. MSA/PCA split method Quick tutorial This is a highly abbreviated form of the full single particle reconstruction tutorial. The numbering follows the detailed tutorial above, so you can switch back and forth. While this quick tutorial will get you through the process, you may not learn much on the way. It is a good starting point for those who already have EMAN2 experience and are just trying to learn what recent changes have been made. IF YOU ENCOUNTER ANY UNEXPECTED BEHAVIOR HERE, PLEASE CHECK THE FULL TUTORIAL ABOVE BEFORE ASKING FOR HELP! Project set up: unzip e2spt_tutorial.zip cd e2spt_tutorial/ribo-10064 Run e2projectmanager.py Select “Tomo” in the “Workflow Mode” dropdown menu Importing raw data: Double click “Raw data” in the workflow menu and select “import tiltseries” Click “Browse” and select all tiltseries within the “data” folder Type “2.16” in the apix input box, check the “import_tiltseries” box, and choose whether to copy, move, or link the raw data into this project. Click “Launch” Automated 3D reconstruction In the workflow menu, double click “3D Reconstruction” and select “Reconstruct tomograms” Click “Browse” at the top right of the menu or select “alltiltseries” at the lower right before “Options” In the “tltstep” box, type “2.0”, and in the “npk” box, type “20”. Click “Launch” Visualize reconstructions (Optional) In the workflow meu, double click “Analysis and Visualization” and select “Evaluate Tomograms” Click “Launch” Reference-based subtomogram boxing (recommended for tutorial) Under the “Subtomogram Averaging” entry in the workflow menu, click “Reference-based boxing”. the provided reference, boxref.hdf On the “tomograms” row, click “Browse” In the browser window, select all reconstructed tomograms and click “OK” On the “reference” row, click “Browse” Select “boxref.hdf” and click “OK”. This file contains a low-resolution reference structure that we will use to box ribosome particles. In the “label” box, type “Ribosomes”. Click “Launch” to begin reference-based boxing. Manual subtomogram boxing (optional) In the workflow menu, double click “Subtomogram Averaging” and select “Manual boxing” Click “Browse” on the right and select the first tiltseries in the project. Click “Launch”. This will open 3 new GUI windows. First, in the “Options” window under “Sets”, click “New” to create a new set of particles. Label this set “Ribosomes” Inside the Sets table, click on the new set labeled “XX :: Ribosomes”, where XX may be “01”, “02”, etc depending on how many particle types are present. In the “Radius” blank at the top of the “Options” GUI widget, type 64, which will set the size of the boxes used when extracting ribosome particles. Next, click on the main boxer window displaying Z-slices in the large window and XZ and YZ slices along the lower and lefthand rows respectively. Using the slider bar at the lower right, pan through the Z-slices of the reconstruction. When you see a particle, decide which slice corresponds (roughly) to its center and click on it in the window. This will create a box colored according to the “XX :: Ribosomes” label displayed in the “Options” window and also populate the “Particles List” window. If a box is created by mistake, it can easily be erased by holding the “Shift” key and clicking on the box. Continue this process until you have boxed at least 50 ribosome particles. Select the particle list window and verify that all boxed particles correspond to ribosomes. If a non-Ribosome particle is observed, hold the Shift key and click on it to erase that box. Once finished, close the main GUI window. Repeat these steps for each tiltseries in the project. Remove bad boxes In the workflow menu under “Analysis and Visualization”, click “Evaluate tomograms” For each reconstructed tomogram, select it in the main window and click “Boxer” to open the subtmogram boxing widget (e2spt_boxer22.py) Scan through the particles shown in the “Particles List” window If a non-ribosome particle is observed, hold the Shift key and click on the particle to remove the box. Close the boxer window and repeat on all tomograms containing boxed ribosome particles. CTF Correction In the workflow menu under “Subtomogram Averaging”, click “CTF correction” Click “Browse”, select every tiltseries, and click “OK” In the dfrange box, type “2.,7.,0.1”. In the voltage box, type “200” In the cs box, type “2.0” Click “Launch” Extract particles In the workflow menu under “Subtomogram Averaging”, click “Extract particles” Click “Browse”, select all tomograms, and click “OK” In the “threads” box, specify the number of cores to use when running this process. Check the box next to “wiener” to turn on Wiener filtering Click “Launch” Build sets In the workflow menu under “Subtomogram Averaging”, click “Build sets” Click “Browse”, select all particle stacks, and check the “allparticles” box. Click “Launch” Generate reference-free initial model In the workflow menu under “Subtomogram Averaging”, click “Generate initial model” Click “Browse”, select the “Ribosome.lst” set, and click “OK” Click “Launch” to begin reference-free initial modeling. 3D refinement In the workflow menu under “Subtomogram Averaging”, click “3D refinement” Next to the particles file box, click “Browse”, select the “Ribosome.lst” particle set, and click “OK” Next to the reference file box, click “Browse”, select the initial model you generated previously, and click “OK” In the “niter” box, type “4” In the “sym” box, type “c1” In the “mass” box, type “3200” In the “tarres” box, type “10” In the “threads” box, specify the number of cores to use when running this process Click “Launch” to begin iterative 3D subtomogram alignment Visualize and analyze refinements In the workflow menu under “Analysis and Visualization”, click “Evaluate SPT refinements” Click “Launch” and examine your last SPT refinement. Sub-tilt refinement In the workflow menu under “Analysis and Visualization”, click “Sub-tilt refinement” Specify the path to the “spt_XX” directory, corresponding to the final 3D refinement In the “iter” box, type 3 to run for 3 iterations. In the “threads” box, specify the number of cores to use when running this process Check the “dopostp” box Click “Launch” Addressing heterogeneity In the workflow menu under “Analysis and Visualization”, click “Multi-reference refinement” Next to the “particles” box, click “Browse” Select the set containing your ribosome particles (“Ribosome.lst”) and click “OK” Specify reference maps corresponding to the various states you hope to draw out of the data If performing a focused classification of particles, specify a mask in the “mask” file box. Use “Browse” to search for this file. In the “threads” box, specify the number of cores to use when running this process In the “tarres” box, specify the resolution target for multi-model refinement. In the “mass” box, type “3200”. Click “Launch” Repeat any of these steps as necessary. Analysis and visualization can be performed between any of the steps mentioned above. |
This tool helps visualize and compare results from multiple subtomogram refinement runs. Launch it from '''Analysis and visualization -> Evaluate SPT refinement'''. In the GUI, you can look at all '''spt_XX''' and '''sptsgd_XX''' folders and compare their options and resulting maps. Switch between folder type using the menu at top right. Click the header of a column to sort the table by its content. Uncheck items in the list at bottom-right to hide corresponding columns. Clicking '''!ShowBrowser''' will bring up the '''[[http://blake.bcm.edu/emanwiki/EMAN2/Programs/e2display|e2display.py]]''' browser in the folder of the selected row. '''!PlotParams''' will plot the Euler angle distribution and other alignment parameters. The 8 columns in the plot are three Euler angles (az, alt, phi), translation in x,y,z, alignment score, and missing wedge coverage score. '''PlotFSCs''' will plot the FSC curve under tight mask from each iteration. |
EMAN2 Tomography Workflow Tutorial
- This tutorial is best suited for EMAN2 built after 09/27/2018. Not everything described in the tutorial was functioning yet in the 2.22 release.
This tutorial uses data from EMPIAR: EMPIAR 10064 (only the 4 mixed CTEM tilt series)
- Time estimates for each step are for a well-configured tomography workstation with a high-speed disk, 64+ GB of RAM and 16+ cores.
- The pixel size in the header of the files are incorrect as provided by EMPIAR. The correct Apix value (2.62) should be specified when importing the images.
Prepare input files (~2 minutes)
- Make a new empty folder for the project and 'cd' into that folder
- Make sure any EMAN2 commands you run are executed from within this folder (not any subfolder)
- You may use "Edit Project" from the Project menu to set default values for the project. While not required, it reduces later errors.
- Make sure the workflow mode is set to "TOMO" not "SPR"
Import tilt series using Raw Data -> Import tilt series
Select the files, and make sure importation says copy
In this step you should enter the correct A/pix for your data in the apix box. For EMPIAR10064, this is 2.62. For your own data, you need to know this number. In later steps you should be able to use -1 (default) for apix.
If your tilt series isn't a single stack file, but is many individual images instead, you will need to use Generate tiltseries to build an image stack. This is not necessary for the tutorial data.
Once the options are set, press Launch
It is critical that the filenames for your data not contain any spaces (replace with underscore) or periods (other than the final period used for the file extension). "" (double underscore) is also reserved for describing modified versions of the same file, and should not be used in your original files.
Tiltseries Alignment and Tomogram Reconstruction (20 min)
Alignment of the tilt-series is performed iteratively in conjunction with tomogram reconstruction. Tomograms are not normally reconstructed at full resolution, generally limited to 1k x 1k or 2k x 2k, but the tilt-series are aligned at full resolution. For high resolution subtomogram averaging, the raw tilt-series data is used, based on coordinates from particle picking in the downsampled tomograms. On a typical workstation reconstruction takes about 4-5 minutes per tomogram.
For the tutorial tilt-series:
3D Reconstruction -> Reconstruct Tomograms
check alltiltseries
alternatively you can select one or more tilt series from the tiltseries folder
check correctrot
tltstep = 2
clipz = 64
If you wish to look at the intermediate aligned tilt-series and other files, uncheck notmp
- This is not required for the remaining steps in the tutorial, but can be used to help understand how the tomogram alignment works. This requires significant additional disk space. You may consider doing this for only one tomogram.
In each tomorecon_XX folder
landmark_0X.txt has the location of the landmarks (which may be fiducials if present) in each iteration
samples_0X.hdf shows the top and side view of those landmarks
ptclali_0X.hdf has the trace of each landmark throughout the tilt series (they should stay at the center of image all the time if the alignment is good)
tomo_0X.hdf is the reconstruction after each iteration
- Launch
When working with your own data:
Either specify the correct tltstep if the tilt series is in order from one extreme to the other, or specify the name of a rawtlt file (as produced by serialem/IMOD).
While the program can automatically compute the orientation of the tilt axis, it is better to fill in the correct value in tltax since there is a handedness ambiguity in the tomogram if determined automatically.
In most cases, the default npk should be fine. If fiducials are present, it is not necessary to adjust this number to match the number of fiducials. The program will use any high contrast areas it finds as potential landmarks.
bytile should normally be selected, as it will normally produce better quality reconstructions at higher speed. If 2k or larger tomograms are created, memory consumption may be high, and you should check the program output for the anticipated RAM usage.
- The graphical interface only permits 1k or 2k reconstruction sizes. In our experience this is normally sufficient for segmentation or particle picking.
When the sample is thin (purified protein, not cells), it is useful to check correctrot to automatically position tomograms flat in ice
It can also be helpful with thin ice to specify a clipz value to generate thinner tomograms (perhaps 64 or 96 for a 1k tomogram).
CTF Estimation (10 min)
For the tutorial tilt-series:
Subtomogram Averaging -> CTF Correction
check alltiltseries
Double check the voltage and cs
- Launch
When working with your own data:
The first two options, dfrange and psrange indicate the defocus and phase shift range to search. They take the format of “start, end, step”, so “2, 5, .1” will search defocus from 2 to 5 um with a step size of 0.1. Units for phase shift is degrees.
For images taken with volta phase plate, we usually have dfrange of “0.2,2,0.1” and psrange of “60,120,2”.
Note that this program is only estimating CTF parameters, taking tilt into account. It is not performing any phase-flipping corrections on whole tomograms. CTF correction is performed later as a per-particle process. This process requires metadata determined during tilt-series alignment, so it cannot be used with tomograms reconstructed using other software packages.
Tomogram annotation (optional)
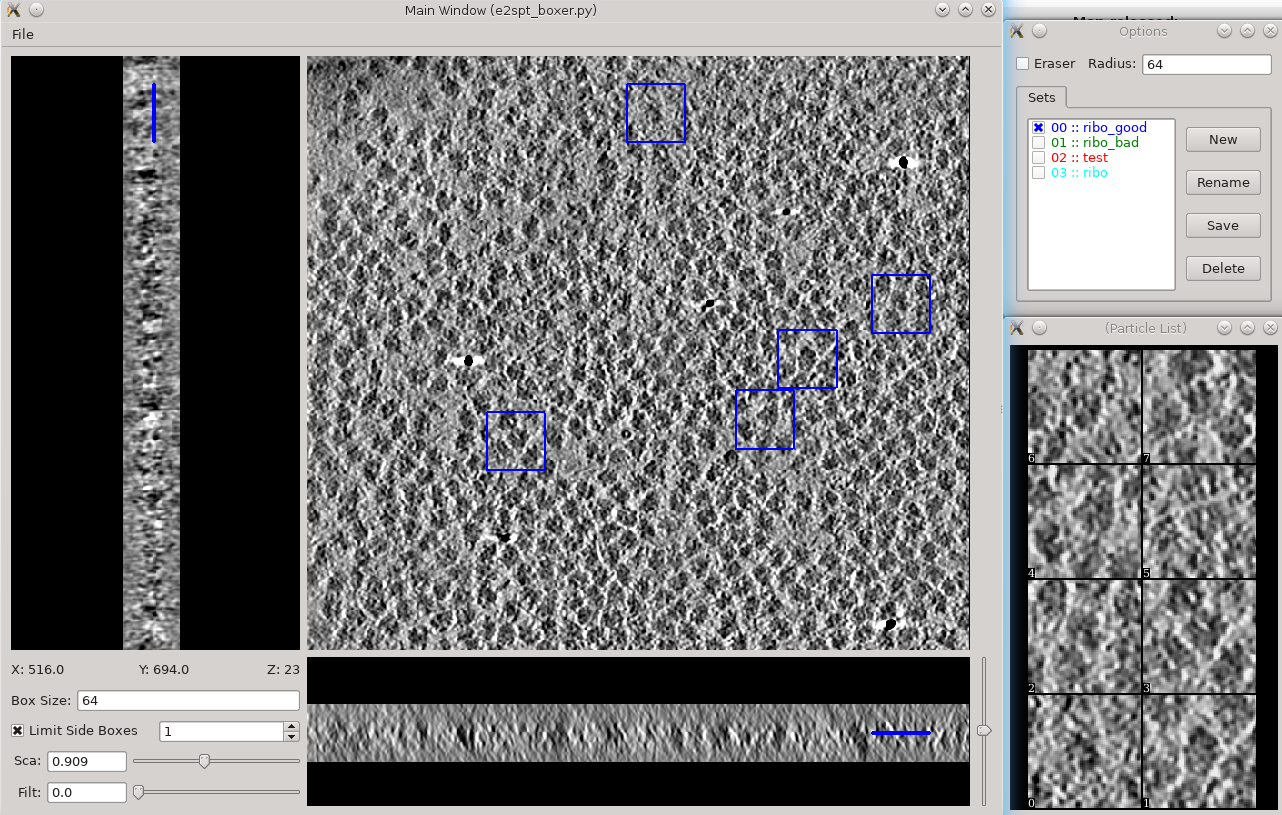
- Since the tutorial data set is purified ribosomes, this step can be skipped for the tutorial data, and you can move on to template-based particle picking. For cells or other types of complex specimens, tomogram annotation can be used to produce locations of different types of objects.
This section is brief and is probably only useful for users already somewhat familiar with the tomogram annotation protocol. A more detailed tutorial of the subject can be found in TomoSeg. Note that some directory structure and user interfaces have changed in the latest version to match new tomogram workflow.
Segmentation -> Preprocess tomograms
- This step is not always necessary for tomograms reconstructed in EMAN2, but may slightly improve results.
Segmentation -> Box Training References
- This is a newer interface than previously used for this step. Select a few "Good" (regions containing the feature of interest) and "Bad" (regions not containing the feature of interest) boxes.
- "~" and "1" on the keyboard can be used to move along the Z axis.
- The new interface permits different types of features to be identified in a single session and in the same tomogram.
If the different features of interest have very different scale, it is always better to keep the box size at 64, and instead rescale the tomogram. As long as the rescaling is done using EMAN2 utilities, the program will correctly keep track of the geometry relative to the original tomogram & tilt series.
- if you are doing this with the tutorial data, you would only have 2 classes of particles "ribo_good" and "ribo_bad".
When pressing Save all visible particles (box checked next to the class name) will be saved
The rest of the annotation process remain unchanged from the original tutorial, except that now, all trained neural networks and training results are saved in the neuralnets folder, and all segmented maps are in the segmentations folder. You now only specify the label of the output file instead of the full file name.
Segmentation -> Find particles from segmentation to turn segmented maps into particle coordinates.
- Input both the tomogram and its corresponding segmentation, and the particles coordinates will be written into the metadata file.
- Slightly tweaking the threshold parameters may yield better results.
featurename will become the label of particles generated. Those particles can be viewed in the particle picking step and processed in the following protocols.
Particle picking
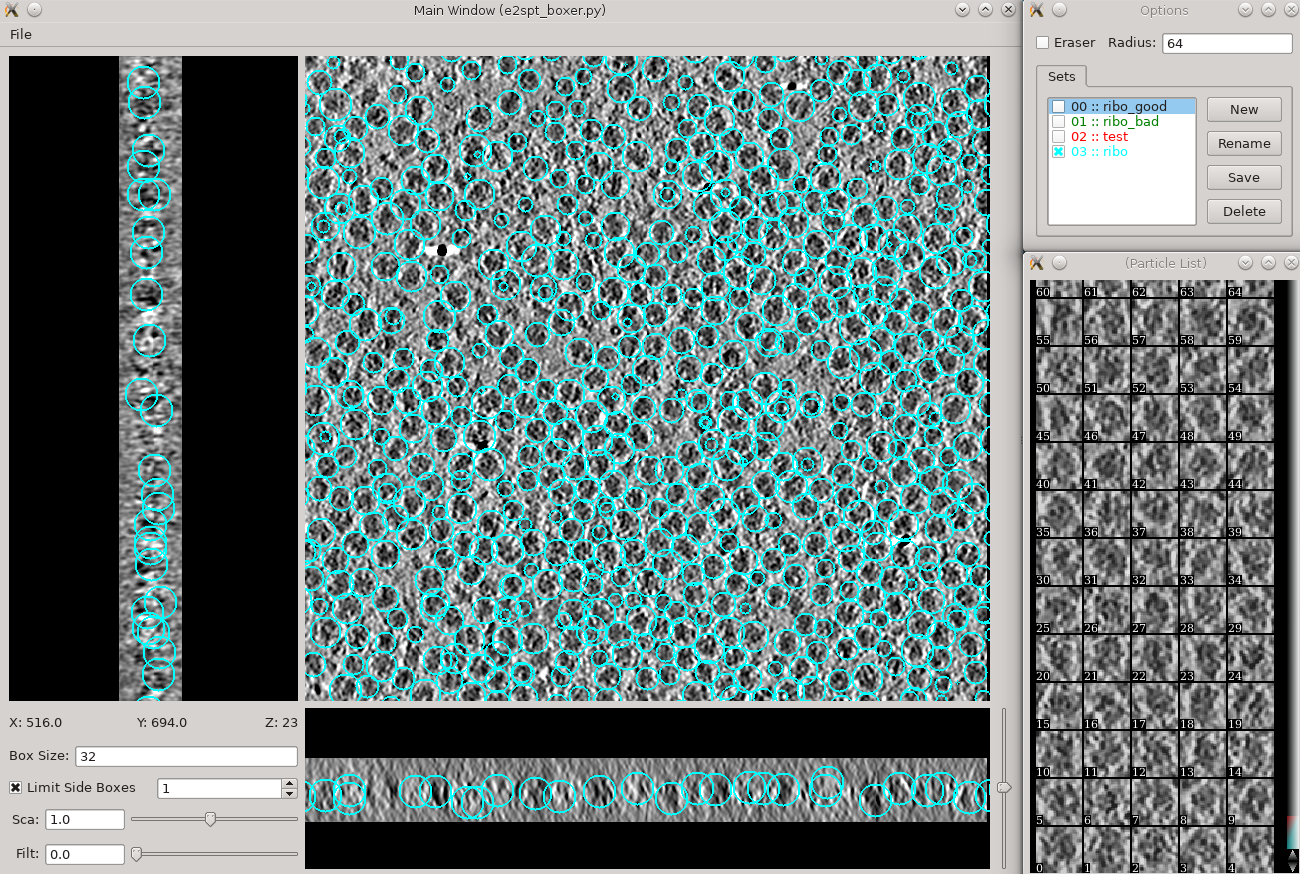
Launch the boxer in Subtomogram averaging -> Manual boxing step. You can also launch it via the Tomogram evaluation step which is discussed later in this tutorial. Go through slices along z-axis using ‘~’ and ‘1’ on the keyboard and deleting boxes by holding Shift and click. The boxes are shown as circles, whose radii indicate the distance from the current slice to the center of the particles. You can rename the label of the boxes and create new types of boxes in the options window. Make sure the label is consistent for the same particle type in different tomograms, as it is used for particle extraction later. The box size can be set in the main window at the left bottom corner, in this case, 45 for the ribosomes (so the unbinned box size is 180). If you have got the particle coordinates from tomogram annotation, just take a look at the automatically generated particles here and remove some obvious bad ones. While you can save 3D particles from the GUI, there is no need to do so in this step. When you are satisfied with the result, simply close the window. You should have ~3000 particles from the 4 tomograms in the dataset.
If you skipped the tomogram annotation step, we will pick a few particles here to generate an initial model first, and use the initial model as a reference for template matching. Select 30-50 particles from a tomogram, then close the boxer window.
Particle extraction
In this step, the program will extract unbinned 2D particles from tilt series, perform per particle per tilt CTF correction, then reconstruct individual 3D particles. Select Extract particles from the left panel, check alltomograms, and specify the label of particle you want to extract. Make sure the label specified here corresponds to the label of particles from the particle boxer. If the box size is correct when you select particles from the GUI, you can leave boxsz_unbin as -1, so the program will keep that box size. You can adjust the value if you want to change the box size of the extracted particles. If your particles are deeply buried in other densities, using a bigger padtwod may help, but doing so may significantly increase the memory usage and slow down the process. With CTF information present, it generally does not hurt to check wiener, which filters the 2D particles by SSNR before reconstructing them in 3D. If you want to generate particles without CTF correction, check noctf. By default, the generated particles will have the same label as they are named in the boxer. If you want to have multiple types of particles, for example, with and without CTF correction, you can specify a different newlabel each time you launch the program. Specify a binning factor in shrink to produce downsampled particles if your memory/storage/CPU time is limited, but it may also limit the resolution you achieve at the end.
For the EMPIAR example, check alltomograms, and specify the label for the particles (either all ribosome particles or the ones for initial model); click Launch.
Then, go to Build set in the left panel, check allparticles, and click launch. This will generate particle sets, which are virtual particle stacks that consist of particles with the same label from different tomograms.
Initial model generation
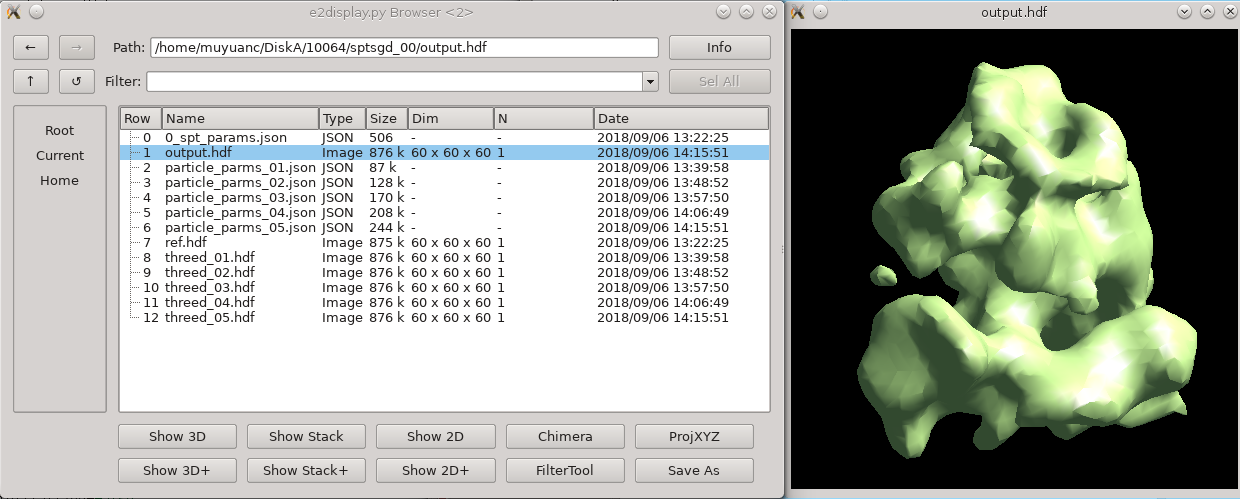
To build an initial model from scratch, simply go to the Generate initial model step and input the particle list. If you wish the process to be faster, set shrink to 2-4. It is not necessary to change other options. The program is parallelized, but not in a standard EMAN2 way. To use more cores, you can enter a bigger number in batchsize. This will not make the program run faster but may make it converge to the correct answer faster. Also using more particles as input won’t make it run faster or slower either, so either input the full particle set if you have them, or the 30-50 particles you pick for the initial model. If the protein is known to be symmetrical, specify the correct symmetry. The program will not actually apply the symmetry (unless you check the applysym box, which is not recommended in general), but it will align the initial model to the symmetry axis so the following steps can work. For most situations, the default number of iterations (niter) of 5 is much more than needed.
In this ribosome dataset with shrink 3, the program should converge to a good initial model before the end of the first iteration, usually within 10 minutes. Output files are written in folders called sptsgd_XX. In the output folder, the file output.hdf is the current initial model, which is updated after each batch (so 10-20 times per iteration). So it is okay to stop the program early and use the file as an initial model once it looks good enough. While it would be good to have a better early stopping criterion, given the diversity of things in the cell, we have not come up with one yet.
Template matching
If you generated all particles with tomogram annotation already, skip this step. If not, go to Reference-based boxing, click Browse for tomograms to select all tomograms, and Browse the initial model generated in the previous step as reference. Specify the label of the output particles in label and set a maximum particle number per tomogram in nptcl (in the EMPIAR example, 800 should be fine), and click Launch.
After the program finishes, take a look at the particle coordinates from Manual boxing in the project manager or Tomogram evaluation, and manually remove the obvious bad boxes. This may perform worse than the tomogram annotation in excluding ice contamination and fiducial, but should still be fine for purified samples in this example. Once you are satisfied with the boxes, repeat the Particle extraction step using the label for the full particle set to generate all 3D particles.
Subtomogram refinement
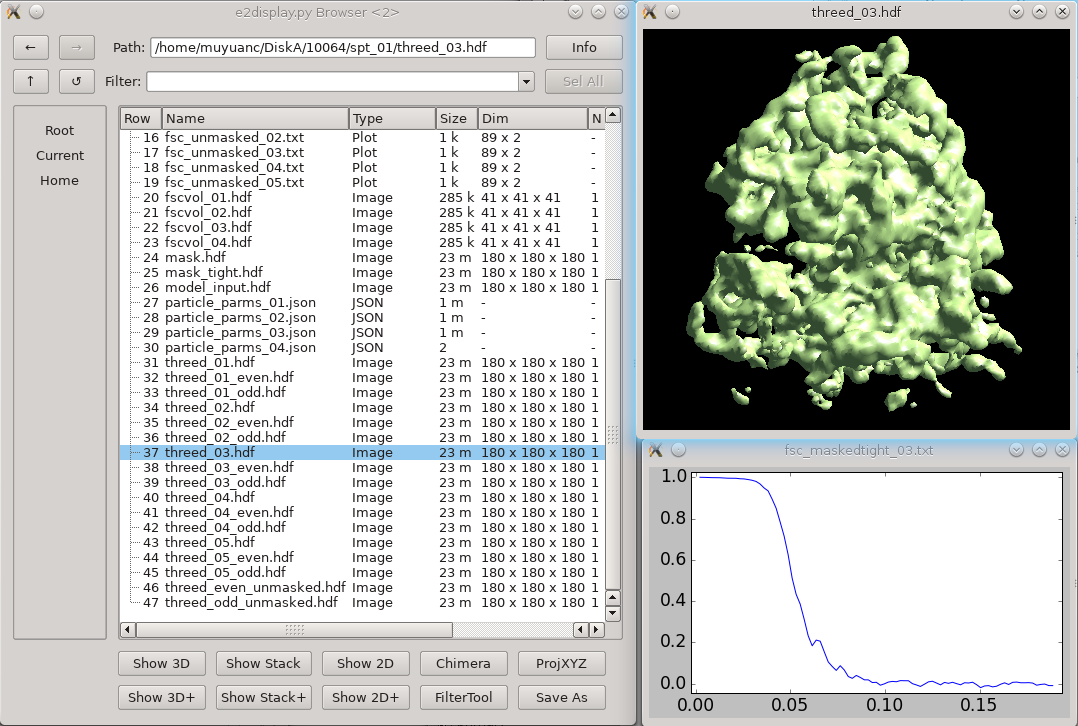
Click 3D refinement from the left panel, and input both the particle set and the initial model generated from the last step as a reference. If there is a symmetry of the protein, make sure it is aligned to the symmetry axis before specifying the correct symmetry. If you are willing to split the even/odd set of particles and do a “gold-standard” refinement, specify a resolution number (usually 30-50) in goldstandard, so information beyond that resolution will be randomized independently in the reference for even and odd set. While it is good to have a reasonable mass for the molecular weight of protein (in kDa) and tarres for the target resolution, leaving them as default usually does not hurt. If you have a known structure factor in a .txt file, (you can compute it from a known structure via e2proc3d.py), specify it in setsf. localfilter will filter the averaged map by local resolution, which is especially useful when looking at things in cells where parts of proteins can be very flexible. This is almost always good to check when you want to push toward high resoluion. pkeep controls the fraction of particles that go into the final average. If you know there are many bad particles in the dataset, setting it to be a smaller number may help. Enter the number of threads you want to use in the thread option. Finally, click Launch and wait. For this dataset, it can take a few hours on a decent workstation. The results can be seen in the spt_XX folder. In the folder, threed_XX.hdf files are the main output map after each iteration, and fsc_masked/unmasked/masktight_XX.txt files are the FSC curves between even/odd half set under different masking. You should be able to get to 12-15Å resolution (cutoff 0.143) at this step using this dataset.
Subtilt refinement
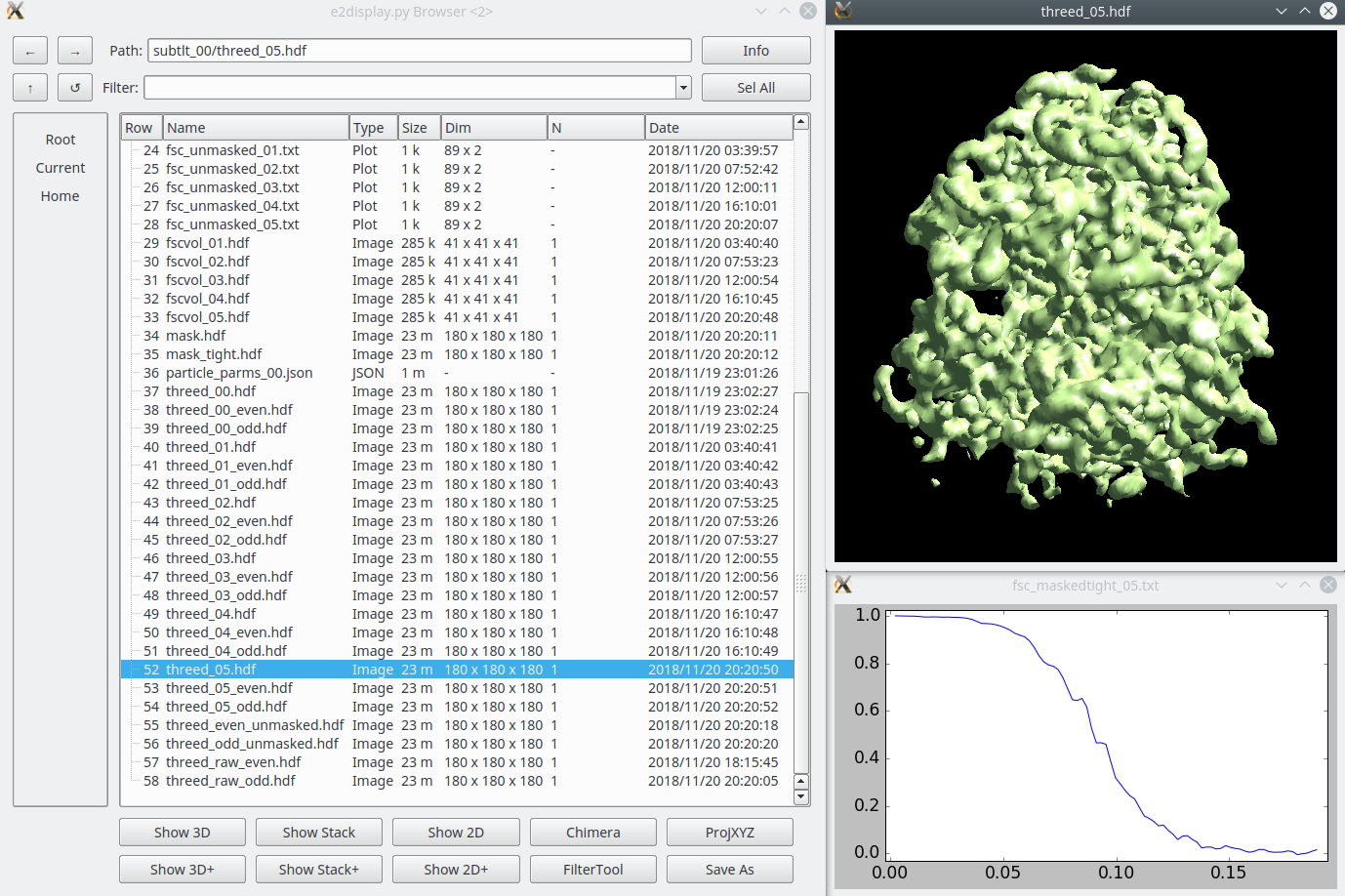
Once the subtomogram refinement finishes, check the final map and FSC curves. In this dataset, you should be able to achieve a resolution of 13-15Å. Now we can refine the orientation of each individual subtilt, i.e. 2D particles from raw tilt series that are reconstructed into to the 3D particles, and push the resolution of the averaged map.
Click Sub-tilt refinement, choose the folder of the last subtomogram refinement and launch the program. You will need to specify the path to the spt_XX directory containing the last completed subtomogram refinement (typically just “spt_00” for example). Additionally, specify the iter you want to use as a starting point for sub-tilt refinement. If “-1” is specified, the program will attempt to locate the last complete iteration.
The default parameters should be generally fine for this dataset, though you may need to alter the parallel and threads options to use the number of CPU threads available on your computer. The niters value corresponds to the number of iterations of sub-tilt refinement you wish to perform. keep controls the fraction of particles that goes into the final map. If you are certain that tilt images beyond a certain angle (for example, 45 degrees) are radiation damaged, you can put 45 in maxalt, and specify a larger keep number. Otherwise, just use keep 0.5, so the program will judge the quality of subtilt images by their correlation to the averaged map and exclude worst 50% 2D particles.
Tomogram evaluation
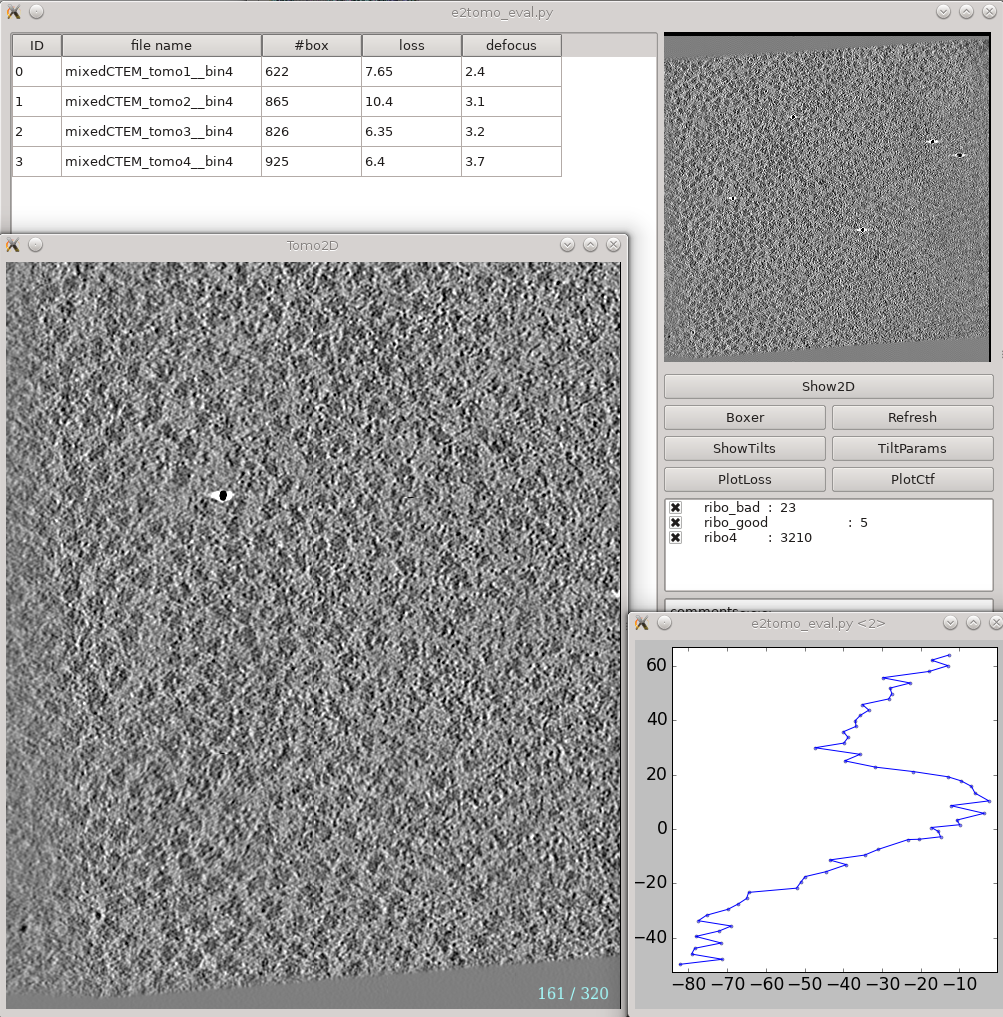
This is a tool that helps you visualize your tomograms with their corresponding metadata, and launch other programs from it. It can be found via Analysis and visualization -> Evaluate tomograms. This can be used at any point of the workflow after tomogram reconstruction.
On the left is a list of tomograms in the project. Clicking the header of each column will sort the table by that attribute. #box is the number of boxes in the tomogram, loss is the average fiducial error in nm, and defocus is the average defocus of the tilt series. Do not be scared by large loss values here. Although the relative value of different tomograms (aligned with the same parameters) in the same project are correlated with tiltseries quality, the exact value here is not as meaningful. You can still get a subnanometer resolution subtomogram average from tilt series with a loss larger than 5 nm.
On the right, the image on the top shows the center slice of the tomogram. The Show2D button shows the selected tomogram in slices, ShowTilts shows the corresponding raw tilt series, and Boxer calls the 3D boxer. PlotLoss will plot the fiducial error per each tilt, and PlotCtf plot the defocus and phase shift at the center of each tilt image. Tiltparams is a bit more complicated. It plots a point list with 6 columns and a number of rows corresponding to the images in the selected tilt series. These are the alignment parameters for the tilt series. The columns represent tilt ID, translation along x and y-axis, tilt angle around y, x and z-axis correspondingly. You can adjust X Col and Y Col in the plot control panel (middle click the plot) to change the display. The first panel below the buttons are the types of particle and their numbers in the dataset. Check and uncheck the boxes will affect the number displayed in #box column on the left. The last box is reserved for comments for each tomogram. You can fill in any comments you have for the selected tomogram and it will be saved with other metadata of the tomogram for future references.
Refinement evaluation
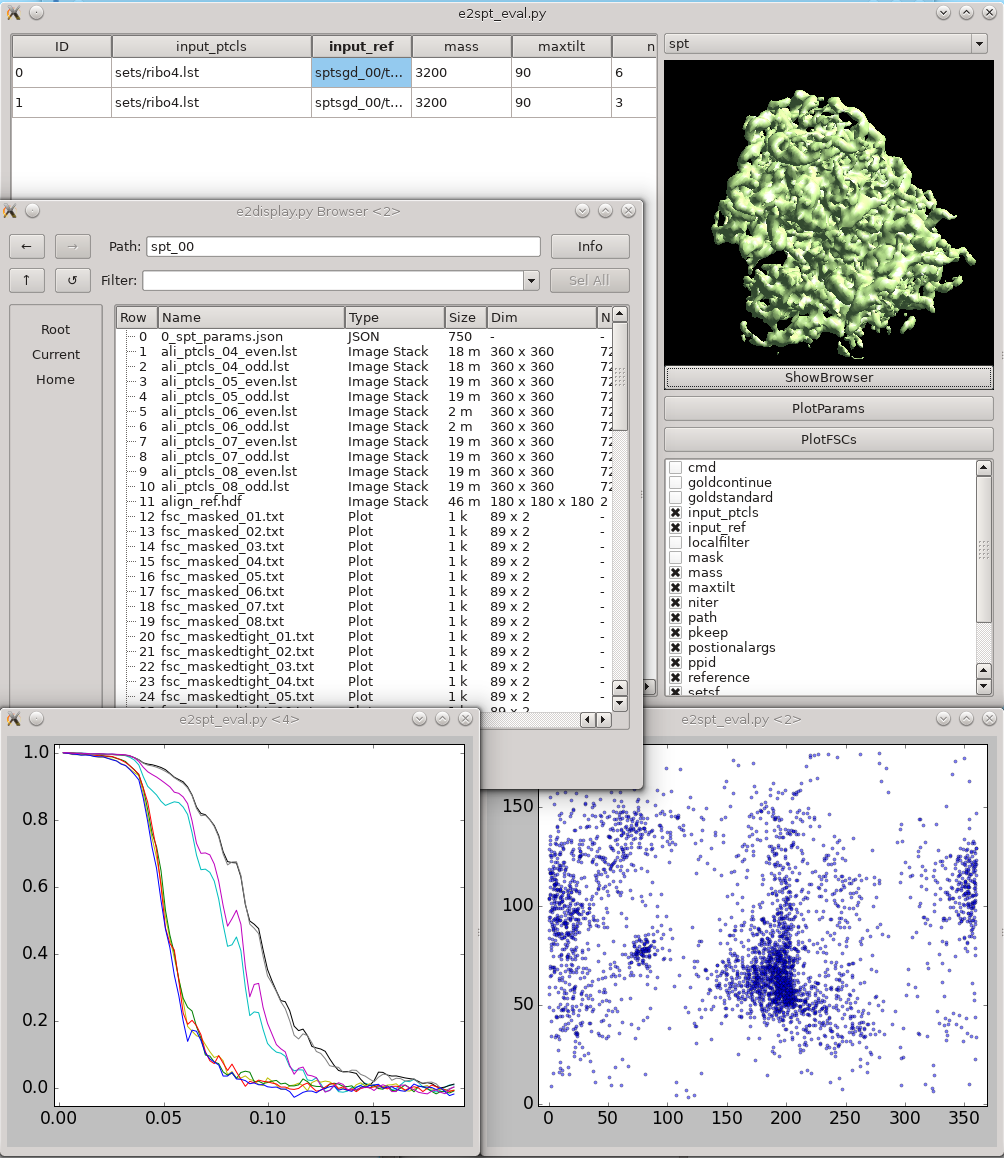
This tool helps visualize and compare results from multiple subtomogram refinement runs. Launch it from Analysis and visualization -> Evaluate SPT refinement. In the GUI, you can look at all spt_XX and sptsgd_XX folders and compare their options and resulting maps. Switch between folder type using the menu at top right. Click the header of a column to sort the table by its content. Uncheck items in the list at bottom-right to hide corresponding columns. Clicking ShowBrowser will bring up the e2display.py browser in the folder of the selected row. PlotParams will plot the Euler angle distribution and other alignment parameters. The 8 columns in the plot are three Euler angles (az, alt, phi), translation in x,y,z, alignment score, and missing wedge coverage score. PlotFSCs will plot the FSC curve under tight mask from each iteration.
