|
Size: 1925
Comment:
|
← Revision 44 as of 2019-04-25 15:01:20 ⇥
Size: 2262
Comment:
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 3: | Line 3: |
| All are welcome to use the materials provided here. Credit is appreciated but not required. | All are welcome to use the materials provided here. Credit is greatly appreciated but not required. |
| Line 9: | Line 9: |
| Note: All protein illustrations in are licensed under a Creative Commons Attribution 4.0 International license by David S. Goodsell and the RCSB PDB. |
|
| Line 15: | Line 13: |
| '''Note:''' The presenter notes in this file contain a full transcription of my talk. |
|
| Line 17: | Line 17: |
| [[attachment:cryoet_poster.pdf]], [[attachment:cryoet_poster.pptx]] | 2019 Sealy Symposium for Structural Biology and Molecular Biophysics & 2018 Keck ARC 2018: [[attachment:cryoet_poster.pptx]] |
| Line 19: | Line 20: |
| [[attachment:scbmb_retreat_2016.pdf]] | Keck Annual Research Conference 2017: [[attachment:keck_arc_2017.pdf]] |
| Line 21: | Line 22: |
| [[attachment:biochem_retreat_2016.pdf]] | Biophysical Society Meeting 2017: [[attachment:biophysical_society_2017.pdf]] |
| Line 23: | Line 24: |
| [[attachment:biophysical_society_2017.pdf]] | 2016 Biochemistry departmental retreat: [[attachment:biochem_retreat_2016.pdf]] |
| Line 25: | Line 26: |
| [[attachment:keck_arc_2017.pdf]] [[attachment:sealy_2018.pdf]] Note, these posters are have been uploaded without modification. There may be typos. |
2016 Structural & Computational Biology and Molecular Biophysics Graduate Program Retreat: [[attachment:scbmb_retreat_2016.pdf]] |
| Line 33: | Line 30: |
| [[attachment:mapchallenge_wrapup_2017.key]] | Slides from the 2015 EMDatabank map/model challenge wrap up meeting: [[attachment:mapchallenge_wrapup_2017.key]] |
| Line 35: | Line 32: |
| [[attachment:microscopy_microanalysis_2018.ppt]] | Slides from the 2018 Microscopy & Microanalysis meeting on the EMAN2 cryoET workflow: [[attachment:microscopy_microanalysis_2018.ppt]] Slides from 2018 Quantitative and Computational Biosciences annual retreat on motion correction: [[attachment:qcbarc2018.key]] |
| Line 39: | Line 38: |
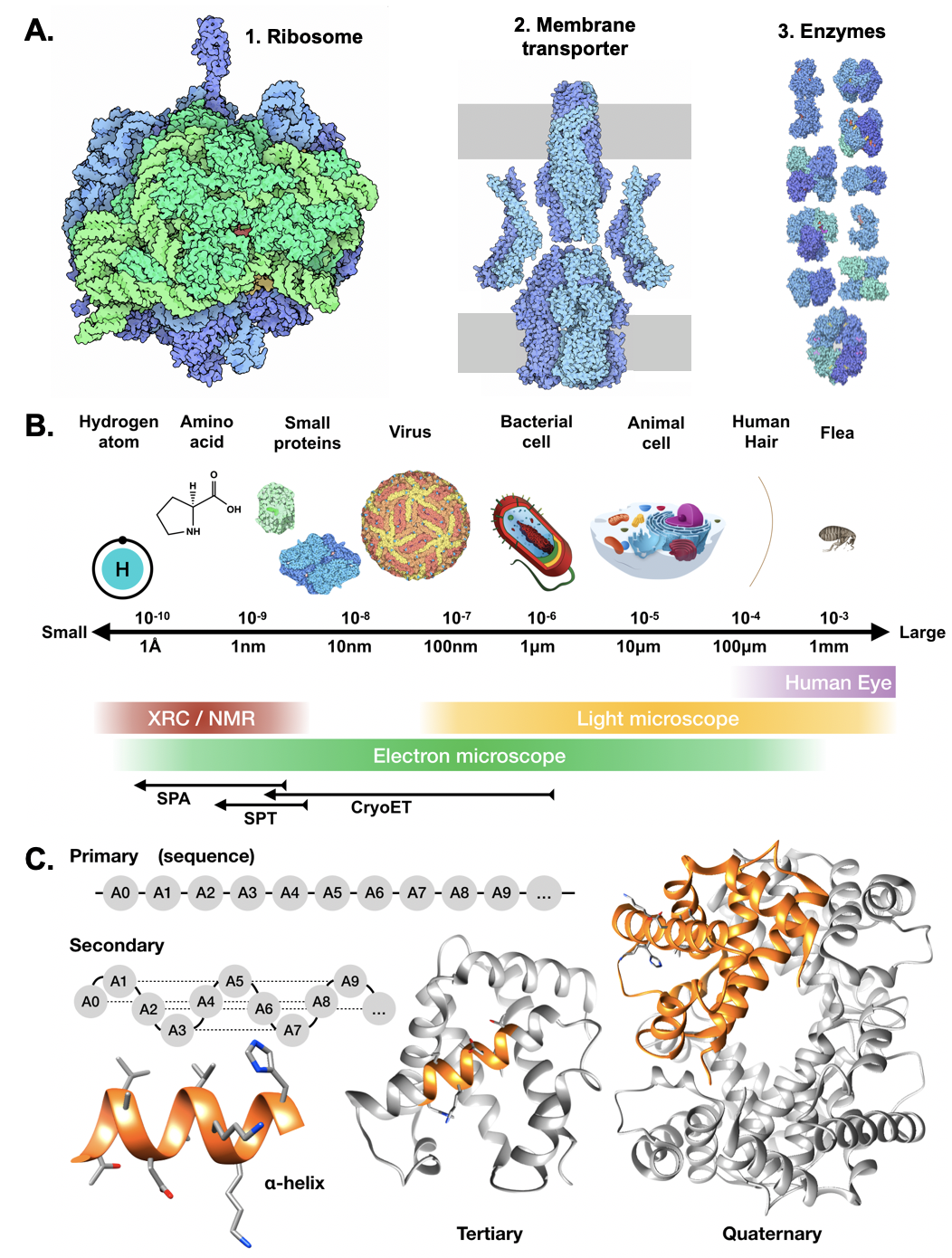
| Structural biology overview {{attachment:struct_bio.png||align="center",width=400}} |
Please see my thesis for corresponding figure captions. |
| Line 42: | Line 40: |
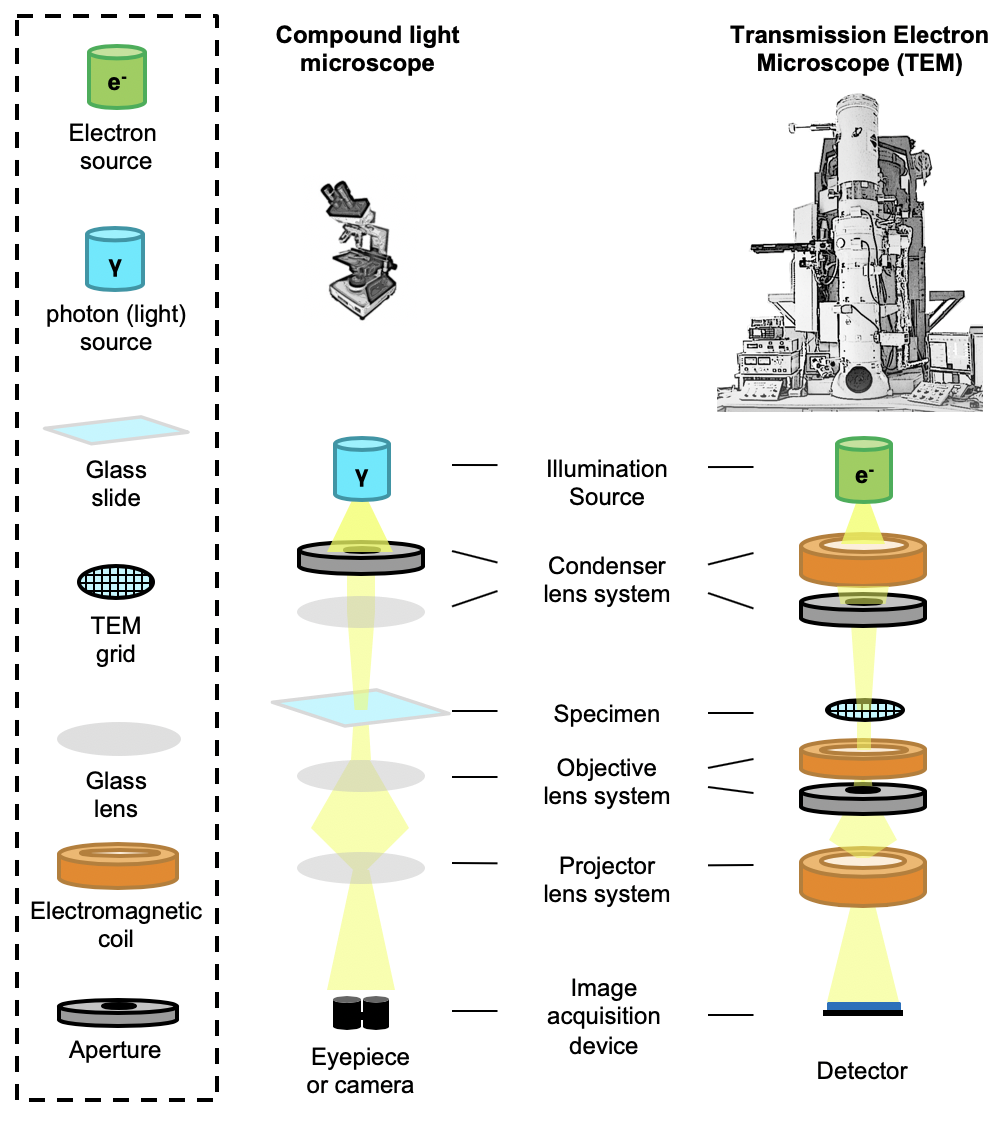
| Transmission electron microscopes (TEM) {{attachment:tem.png||align="center",width=400}} |
|
| Line 45: | Line 41: |
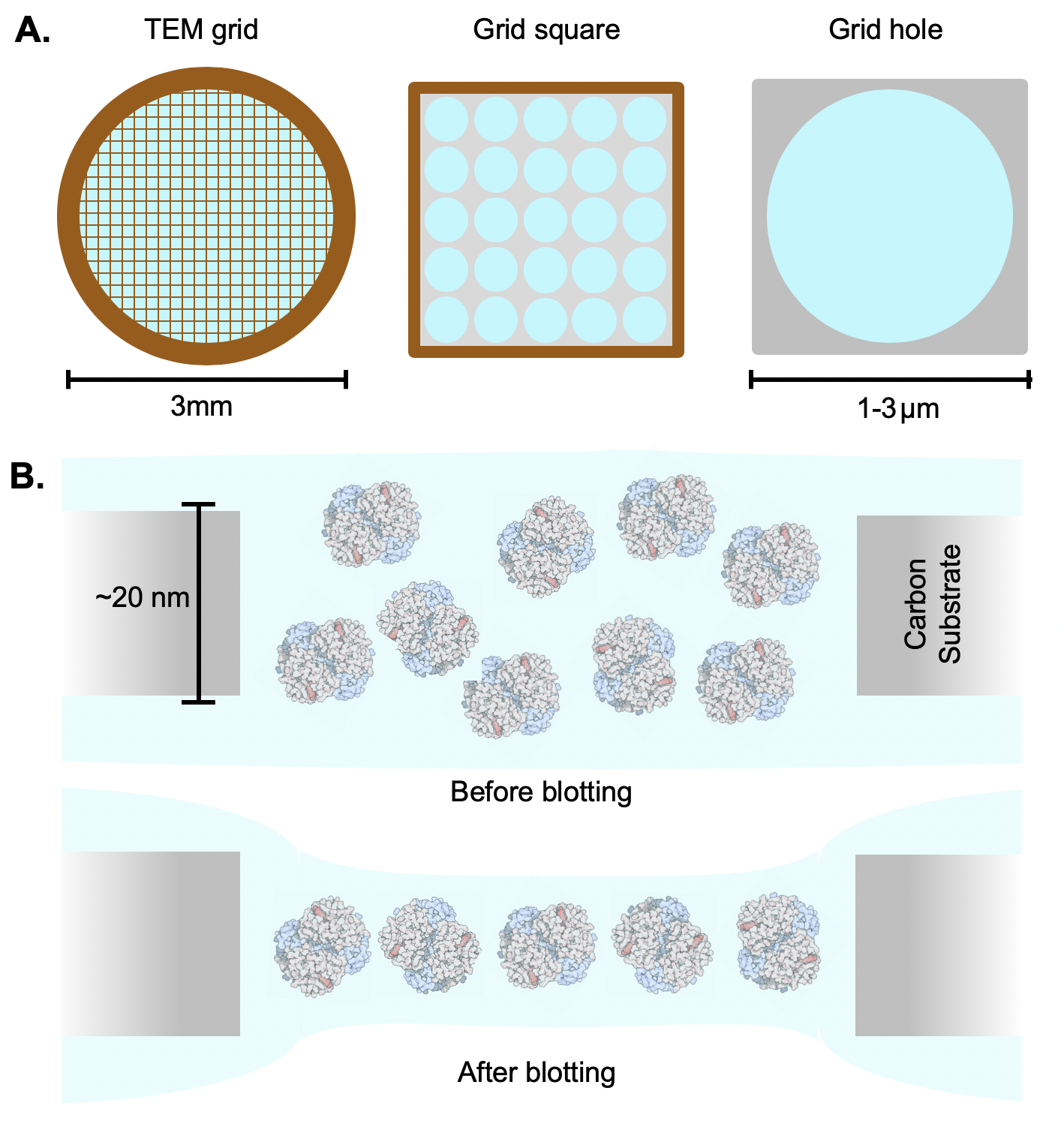
| TEM grids {{attachment:grid.png||align="center",width=400}} |
'''Structural biology overview''' |
| Line 48: | Line 43: |
| Movie-mode imaging {{attachment:movie.png||align="center",width=400}} |
{{attachment:struct_bio.png||width=400}} |
| Line 51: | Line 45: |
| Basic image processing {{attachment:imgproc.png||align="center",width=400}} |
'''Transmission electron microscopes (TEM)''' |
| Line 54: | Line 47: |
| Fourier transforms {{attachment:fft.png||align="center",width=400}} |
{{attachment:tem.png||width=400}} |
| Line 57: | Line 49: |
| Single particle analysis {{attachment:spa.png||align="center",width=400}} |
'''TEM grids''' |
| Line 60: | Line 51: |
| Contrast transfer function (CTF) {{attachment:ctf.png||align="center",width=400}} |
{{attachment:grid.png||width=400}} |
| Line 63: | Line 53: |
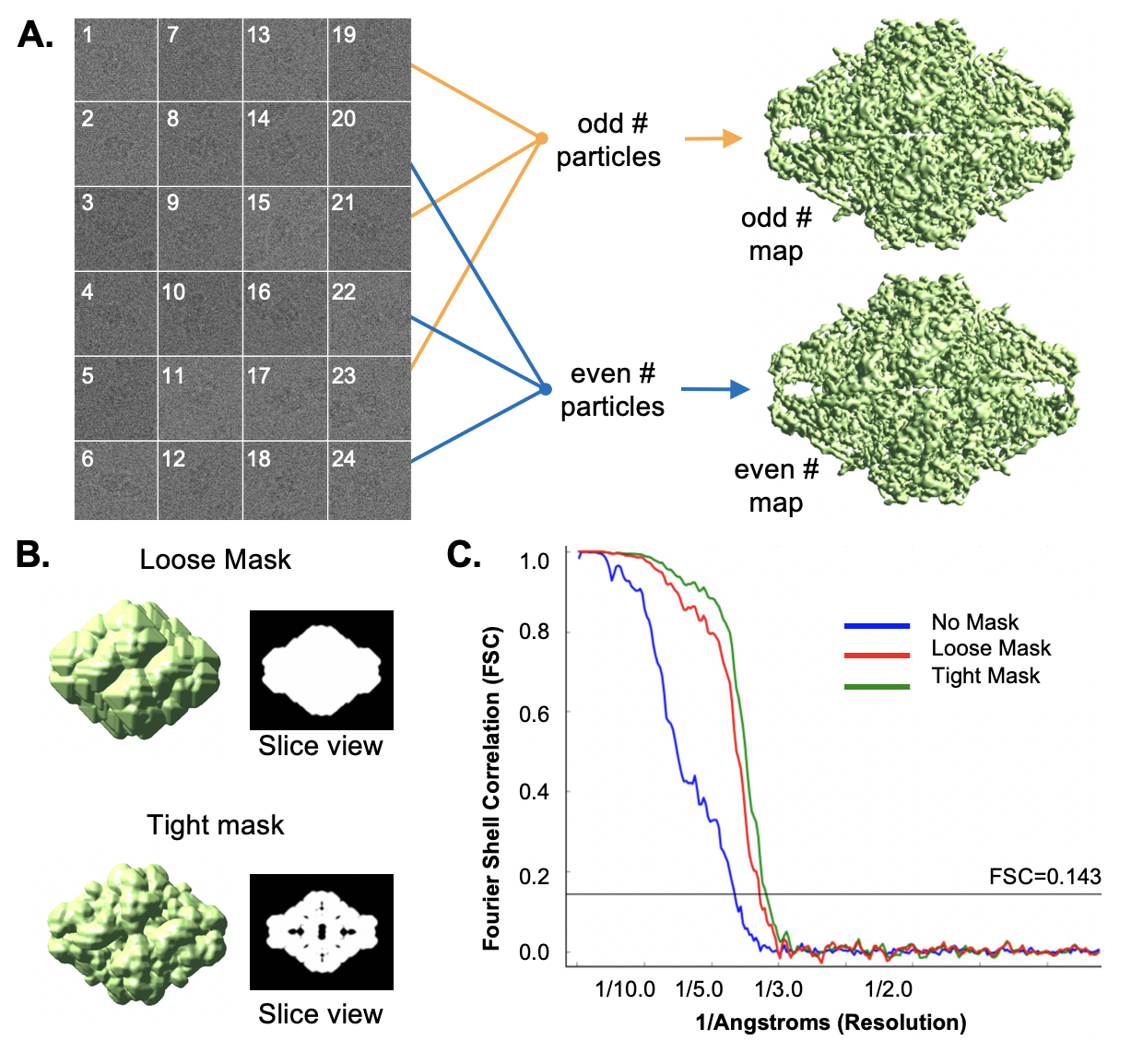
| Resolution measurement via Fourier Shell Correlation (FSC) {{attachment:fsc.png||align="center",width=400}} |
'''Movie-mode imaging''' |
| Line 66: | Line 55: |
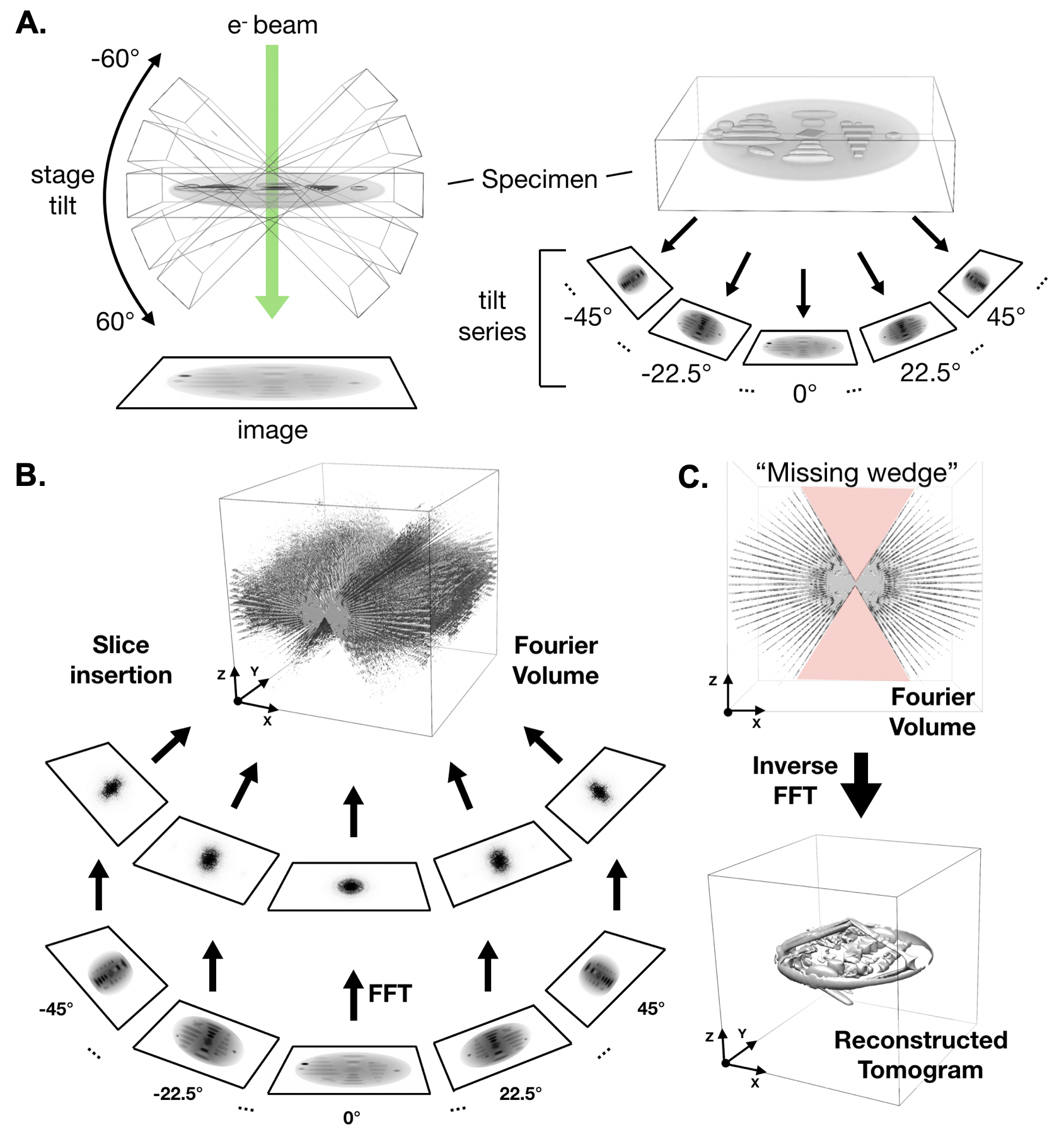
| Electron cryo-tomography (cryoET) {{attachment:cryoet.png||align="center",width=400}} |
{{attachment:movie.png||width=400}} |
| Line 69: | Line 57: |
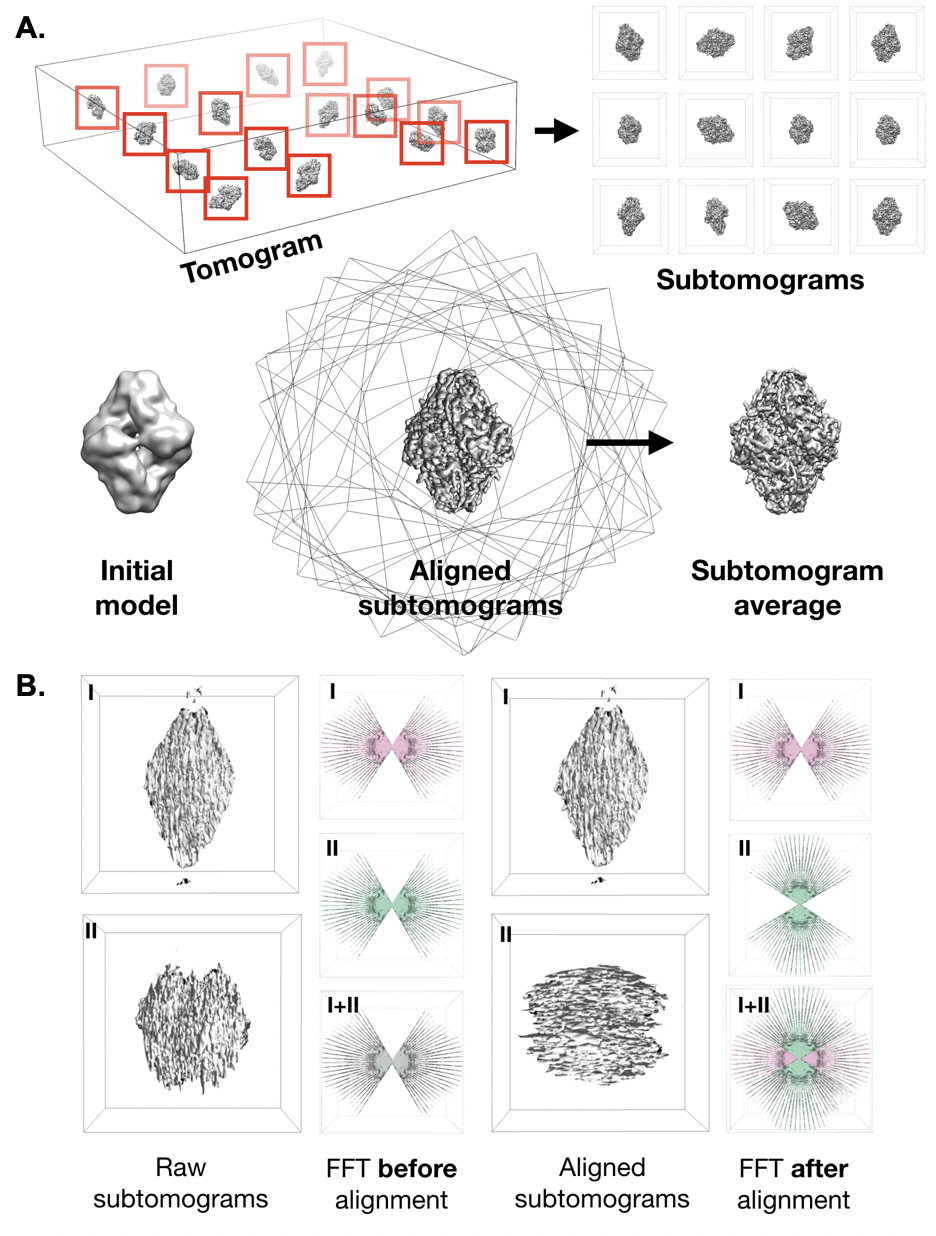
| Subtomogram Averaging / Single particle tomography (SPT) {{attachment:subtomoavg.png||align="center",width=400}} |
'''Basic image processing''' {{attachment:imgproc.png||width=400}} '''Fourier transforms''' {{attachment:fft.png||width=400}} '''Single particle analysis''' {{attachment:spa.png||width=400}} '''Contrast transfer function (CTF)''' {{attachment:ctf.png||width=400}} '''Gold Standard resolution measurement via Fourier Shell Correlation (FSC)''' {{attachment:fsc.png||width=400}} '''Electron cryo-tomography (cryoET)''' {{attachment:cryoet.png||width=400}} '''Subtomogram Averaging''' {{attachment:subtomoavg.png||width=400}} |
Michael Bell, Ludtke Lab (2014 - 2019)
All are welcome to use the materials provided here. Credit is greatly appreciated but not required.
Thesis
Thesis Defense Presentation
Note: The presenter notes in this file contain a full transcription of my talk.
Posters
2019 Sealy Symposium for Structural Biology and Molecular Biophysics & 2018 Keck ARC 2018:
Keck Annual Research Conference 2017: keck_arc_2017.pdf
Biophysical Society Meeting 2017: biophysical_society_2017.pdf
2016 Biochemistry departmental retreat: biochem_retreat_2016.pdf
2016 Structural & Computational Biology and Molecular Biophysics Graduate Program Retreat: scbmb_retreat_2016.pdf
Talks
Slides from the 2015 EMDatabank map/model challenge wrap up meeting: mapchallenge_wrapup_2017.key
Slides from the 2018 Microscopy & Microanalysis meeting on the EMAN2 cryoET workflow: microscopy_microanalysis_2018.ppt
Slides from 2018 Quantitative and Computational Biosciences annual retreat on motion correction: qcbarc2018.key
Select Thesis Figures
Please see my thesis for corresponding figure captions.
Structural biology overview
Transmission electron microscopes (TEM)
TEM grids
Movie-mode imaging
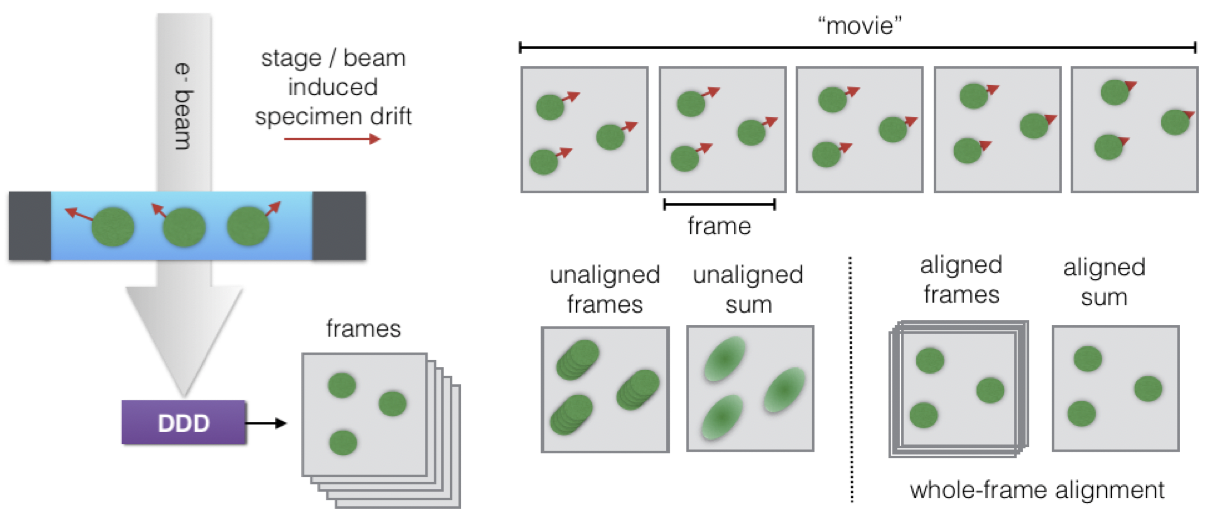
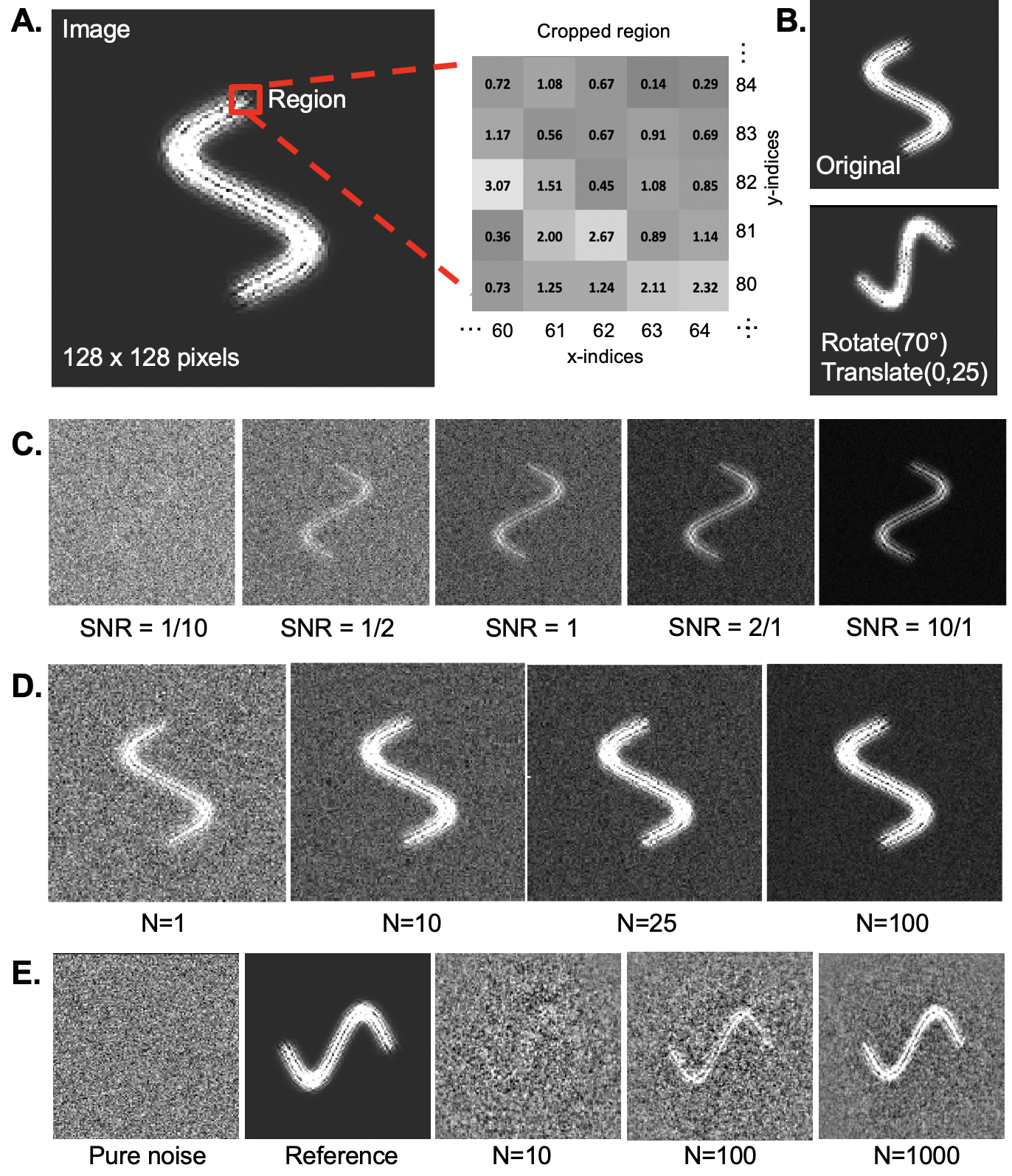
Basic image processing
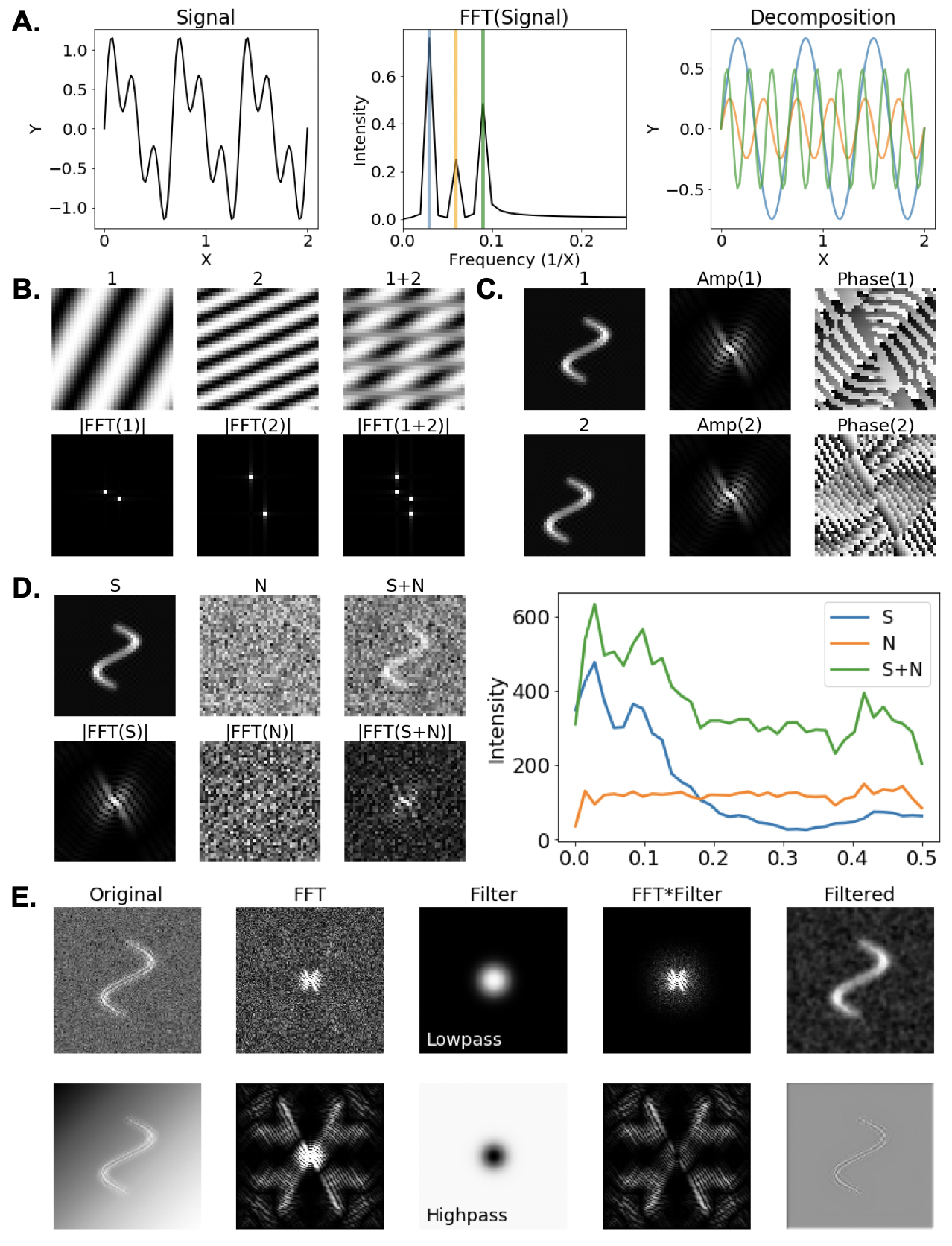
Fourier transforms
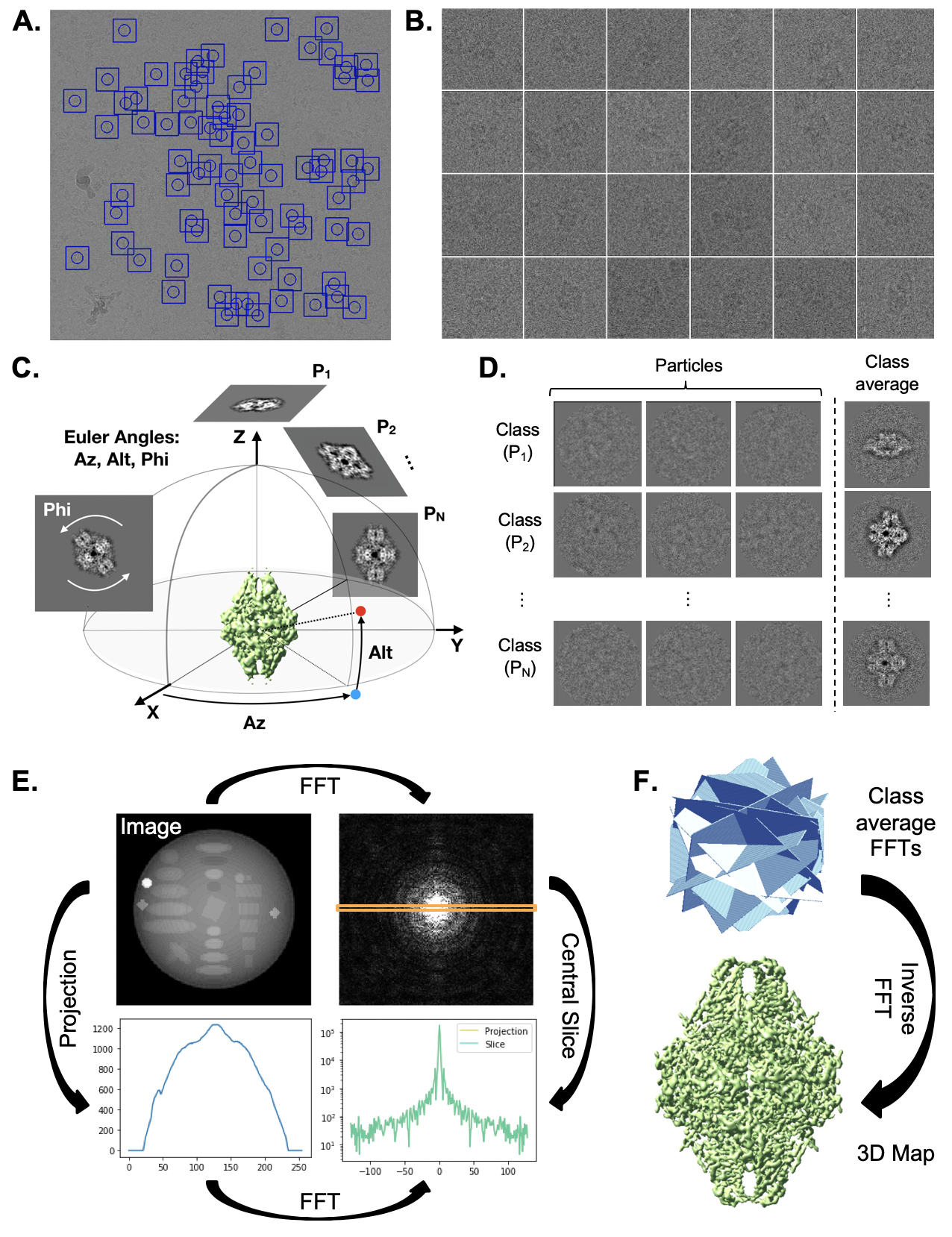
Single particle analysis
Contrast transfer function (CTF)
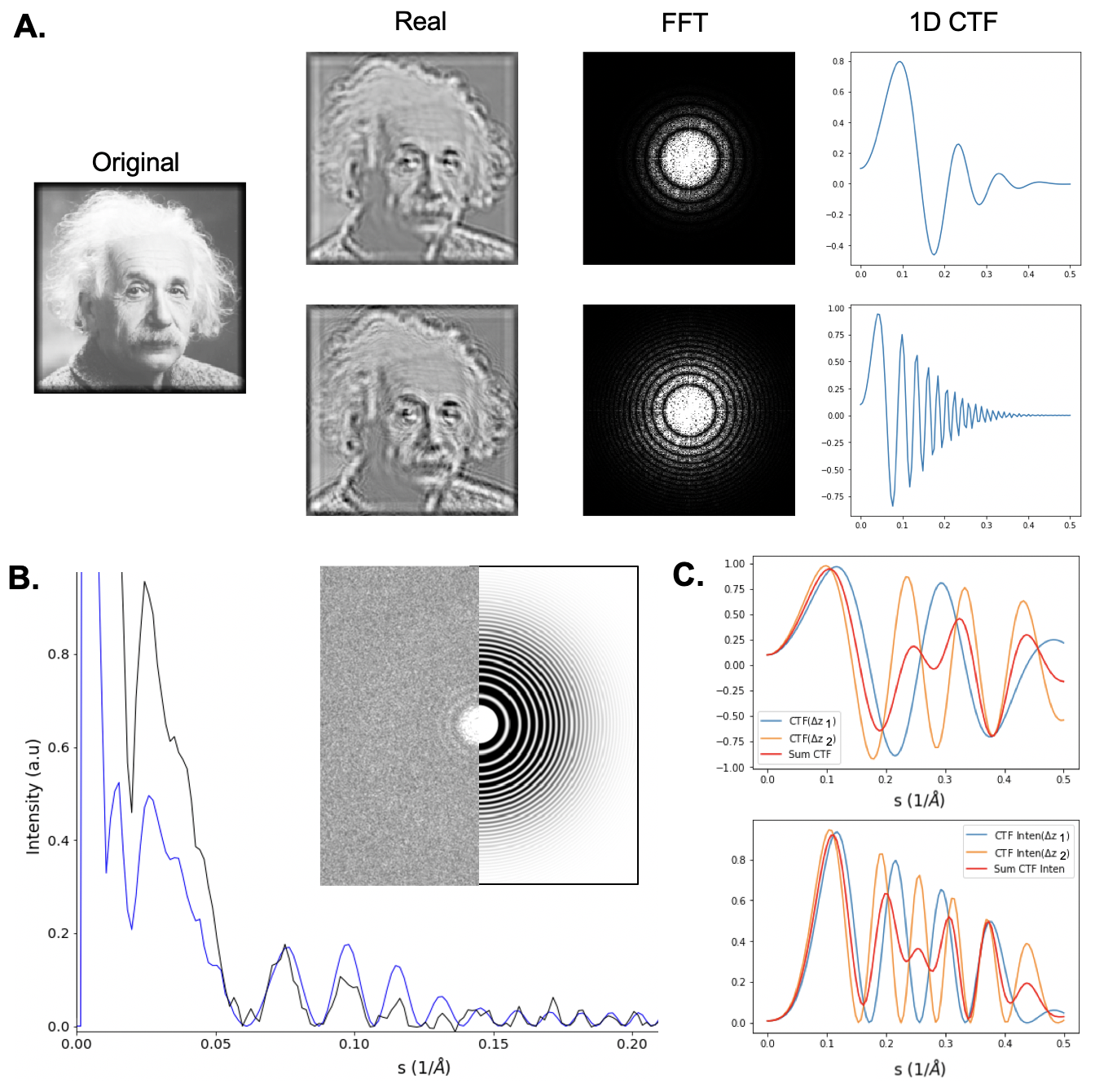
Gold Standard resolution measurement via Fourier Shell Correlation (FSC)
Electron cryo-tomography (cryoET)
Subtomogram Averaging